Splicing Characterization of CLCNKB Variants in Four Patients With Type III Bartter Syndrome
- PMID: 32153641
- PMCID: PMC7047732
- DOI: 10.3389/fgene.2020.00081
Splicing Characterization of CLCNKB Variants in Four Patients With Type III Bartter Syndrome
Abstract
Objective: Type III Bartter syndrome (BS) is caused by loss-of-function mutations in the gene encoding basolateral chloride channel ClC-Kb (CLCNKB), and is characterized by hypokalemic metabolic alkalosis and hyperreninemic hyperaldosteronism. Here, we investigated the molecular defects in four Chinese children with clinical manifestations of Bartter syndrome.
Methods: The genomic DNA of the four patients was screened for gene variations using whole-exome sequencing (WES). The candidate variants were validated by direct Sanger sequencing. Quantitative PCR (qPCR) was subsequently performed to confirm the whole CLCNK gene deletion mutation. A minigene assay and reverse transcription PCR (RT-PCR) were performed to analyze the effect of splice variants in vitro.
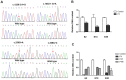
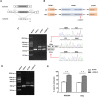
Results: Our patients showed early onset age with hyponatremia, hypokalemia, hypochloremia, repeated vomiting and growth retardation, suggesting Bartter syndrome. Genetic analysis revealed that all patients carried compound heterozygous or homozygous truncating variants in the CLCNKB gene. In particular, we identified a novel nonsense variant c.239G > A (p.(Trp80*)), two splice site variants (c.1053-1 G > A and c.1228-2A > G), a whole gene deletion, and a novel synonymous variant c.228A > C (p.(Arg76Arg)) which located -2 bp from the 5' splice donor site in exon 3. Furthermore, our in vitro minigene analysis revealed c.228A > C, c.1053-1G > A, and c.1228-2A > G cause the skipping of exon 3, exon 12, and exon 13, respectively.
Conclusion: Our results support that the whole CLCNKB gene deletion is the most common mutation in Chinese patients with type III BS, and truncating and whole gene deletion variants may account for a more severe phenotype of patients. We verified the pathogenic effect of three splicing variants (c.228A > C, c.1053-1G > A, and c.1228-2A > G) which disturbed the normal mRNA splicing, suggesting that splice variants play an important role in the molecular basis of type III BS, and careful molecular profiling of these patients will be essential for future effective personalized treatment options.
Keywords: CLCNKB; abnormal RNA splicing; classical Bartter syndrome; hypokalemia; synonymous variant.
Copyright © 2020 Wang, Han, Zhou, Zheng, Zhou, Bao, Jia, Zhang, Huang, Ding and Zhao.
Figures

References
-
- Fraile-Bethencourt E., Diez-Gomez B., Velasquez-Zapata V., Acedo A., Sanz D. J., Velasco E. A. (2017). Functional classification of DNA variants by hybrid minigenes: identification of 30 spliceogenic variants of BRCA2 exons 17 and 18. PloS Genet. 13 (3), e1006691. 10.1371/journal.pgen.1006691 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

