Long Non-coding RNA MIR4435-2HG Promotes Colorectal Cancer Proliferation and Metastasis Through miR-206/YAP1 Axis
- PMID: 32154166
- PMCID: PMC7044350
- DOI: 10.3389/fonc.2020.00160
Long Non-coding RNA MIR4435-2HG Promotes Colorectal Cancer Proliferation and Metastasis Through miR-206/YAP1 Axis
Abstract
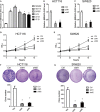
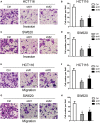
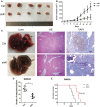
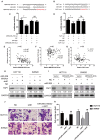
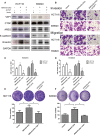
Objective: Long non-coding RNAs (lncRNAs) are critical to colorectal cancer (CRC) progression. In the current study, the objective was the exploration of the role played by lncRNA MIR4435-2HG in CRC proliferation and metastasis. Methods: lncRNA MIR4435-2HG expression and its association with CRC were analyzed using database and clinical specimens. The influences exerted by MIR4435-2HG on cell proliferating process, invading process, and migrating process of CRC were identified after MIR4435-2HG knockdown. The influences exerted by MIR4435-2HG on tumor growth and metastasis were assessed in vivo. The underlying mechanistic associations between MIR4435-2HG, microRNA miR-206, and the transcription factor Yes-associated protein 1 (YAP1) were assessed using bioinformatics and a luciferase reporter gene assay. Results: MIR4435-2HG was highly expressed in CRC tissue in contrast with that in regular tissues and displayed relations to poor prognosis. MIR4435-2HG knockdown could suppress CRC cell proliferation, invasion, and migration. Moreover, MIR4435-2HG knockdown inhibited CRC growth and liver metastasis in vitro. We found MIR4435-2HG knockdown reduced YAP1, CTGF, AREG, vimentin, Snail, Slug, and Twist expression but enhanced E-cadherin expression. Functionally, MIR4435-2HG acted as a competing endogenous RNA (ceRNA) to upregulate YAP1 by sponging miR-206. Conclusions: MIR4435-2HG promoted CRC growth and metastasis through miR-206/YAP1 axis and is likely to play prognostic marker roles and be therapeutically targeted in CRC.
Keywords: MIR4435-2HG; YAP1; colorectal cancer; long non-coding RNA; metastasis; proliferation.
Copyright © 2020 Dong, Yang, Yang, Li and Qiu.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

