Thermal Bioprinting Causes Ample Alterations of Expression of LUCAT1, IL6, CCL26, and NRN1L Genes and Massive Phosphorylation of Critical Oncogenic Drug Resistance Pathways in Breast Cancer Cells
- PMID: 32154227
- PMCID: PMC7047130
- DOI: 10.3389/fbioe.2020.00082
Thermal Bioprinting Causes Ample Alterations of Expression of LUCAT1, IL6, CCL26, and NRN1L Genes and Massive Phosphorylation of Critical Oncogenic Drug Resistance Pathways in Breast Cancer Cells
Abstract
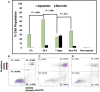
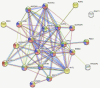
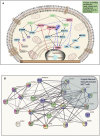
Bioprinting technology merges engineering and biological fields and together, they possess a great translational potential, which can tremendously impact the future of regenerative medicine and drug discovery. However, the molecular effects elicited by thermal inkjet bioprinting in breast cancer cells remains elusive. Previous studies have suggested that bioprinting can be used to model tissues for drug discovery and pharmacology. We report viability, apoptosis, phosphorylation, and RNA sequence analysis of bioprinted MCF7 breast cancer cells at separate timepoints post-bioprinting. An Annexin A5-FITC apoptosis stain was used in combination with flow cytometry at 2 and 24 h post-bioprinting. Antibody arrays using a Human phospho-MAPK array kit was performed 24 h post-bioprinting. RNA sequence analysis was conducted in samples collected at 2, 7, and 24 h post-bioprinting. The post-bioprinting cell viability averages were 77 and 76% at 24 h and 48 h, with 31 and 64% apoptotic cells at 2 and 24 h after bioprinting. A total of 21 kinases were phosphorylated in the bioprinted cells and 9 were phosphorylated in the manually seeded controls. The RNA seq analysis in the bioprinted cells identified a total of 12,235 genes, of which 9.7% were significantly differentially expressed. Using a ±2-fold change as the cutoff, 266 upregulated and 206 downregulated genes were observed in the bioprinted cells, with the following 5 genes uniquely expressed NRN1L, LUCAT1, IL6, CCL26, and LOC401585. This suggests that thermal inkjet bioprinting is stimulating large scale gene alterations that could potentially be utilized for drug discovery. Moreover, bioprinting activates key pathways implicated in drug resistance, cell motility, proliferation, survival, and differentiation.
Keywords: bioprinting; drug discovery; kinase phosphorylation; molecular properties; tumor model.
Copyright © 2020 Campbell, Mohl, Gutierrez, Varela-Ramirez and Boland.
Figures


References
-
- Alvarez E., Northwood I. C., Gonzalez F. A., Latour D. A., Seth A., Abate C., et al. (1991). Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J. Biol. Chem. 266, 15277–15285 - PubMed
-
- Andrade E. L., Bento A. F., Cavalli J., Oliveira S. K., Freitas C. S., Marcon R., et al. (2016). Non-clinical studies required for new drug development-Part I: early in silico and in vitro studies, new target discovery and validation, proof of principles and robustness of animal studies. Brazil. J. Med. Biol. Res. 49:e5644. 10.1590/1414-431x20165644 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

