Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein
- PMID: 32155444
- PMCID: PMC7102599
- DOI: 10.1016/j.cell.2020.02.058
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein
Erratum in
-
Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.Cell. 2020 Dec 10;183(6):1735. doi: 10.1016/j.cell.2020.11.032. Cell. 2020. PMID: 33306958 Free PMC article. No abstract available.
Abstract
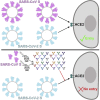
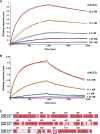
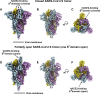
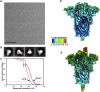
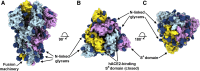
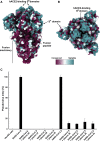
The emergence of SARS-CoV-2 has resulted in >90,000 infections and >3,000 deaths. Coronavirus spike (S) glycoproteins promote entry into cells and are the main target of antibodies. We show that SARS-CoV-2 S uses ACE2 to enter cells and that the receptor-binding domains of SARS-CoV-2 S and SARS-CoV S bind with similar affinities to human ACE2, correlating with the efficient spread of SARS-CoV-2 among humans. We found that the SARS-CoV-2 S glycoprotein harbors a furin cleavage site at the boundary between the S1/S2 subunits, which is processed during biogenesis and sets this virus apart from SARS-CoV and SARS-related CoVs. We determined cryo-EM structures of the SARS-CoV-2 S ectodomain trimer, providing a blueprint for the design of vaccines and inhibitors of viral entry. Finally, we demonstrate that SARS-CoV S murine polyclonal antibodies potently inhibited SARS-CoV-2 S mediated entry into cells, indicating that cross-neutralizing antibodies targeting conserved S epitopes can be elicited upon vaccination.
Keywords: SARS-CoV; SARS-CoV-2; antibodies; coronavirus; cryo-EM; neutralizing antibodies; spike glycoprotein; viral receptor.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing financial interests.
Figures


References
-
- Agirre J., Iglesias-Fernández J., Rovira C., Davies G.J., Wilson K.S., Cowtan K.D. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 2015;22:833–834. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

