Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp
- PMID: 32155802
- PMCID: PMC7157202
- DOI: 10.3390/pathogens9030195
Lineage, Antimicrobial Resistance and Virulence of Citrobacter spp
Abstract
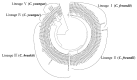
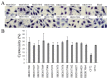
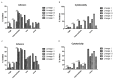
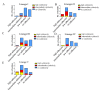
Citrobacter spp. are opportunistic human pathogens which can cause nosocomial infections, sporadic infections and outbreaks. In order to determine the genetic diversity, in vitro virulence properties and antimicrobial resistance profiles of Citrobacter spp., 128 Citrobacter isolates obtained from human diarrheal patients, foods and environment were assessed by multilocus sequence typing (MLST), antimicrobial susceptibility testing and adhesion and cytotoxicity testing to HEp-2 cells. The 128 Citrobacter isolates were typed into 123 sequence types (STs) of which 101 were novel STs, and these STs were divided into five lineages. Lineages I and II contained C. freundii isolates; Lineage III contained all C. braakii isolates, while Lineage IV and V contained C. youngae isolates. Lineages II and V contained more adhesive and cytotoxic isolates than Lineages I, III, and IV. Fifty-one of the 128 isolates were found to be multidrug-resistant (MDR, ≥3) and mainly distributed in Lineages I, II, and III. The prevalence of quinolone resistance varied with Lineage III (C. braakii) having the highest proportion of resistant isolates (52.6%), followed by Lineage I (C. freundii) with 23.7%. Seven qnrB variants, including two new alleles (qnrB93 and qnrB94) were found with Lineage I being the main reservoir. In summary, highly cytotoxic MDR isolates from diarrheal patients may increase the risk of severe disease.
Keywords: Citrobacter; adhesion; cytotoxicity; multidrug resistance; sequence types.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Park Y.-J., Yu J.K., Lee S., Oh E.J., Woo G.-J. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 2007;60:868–871. doi: 10.1093/jac/dkm266. - DOI - PubMed
-
- Tschäpe H., Prager R., Streckel W., Fruth A., Tietze E., Böhme G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: Green butter as the infection source. Epidemiol. Infect. 1995;114:441–450. doi: 10.1017/S0950268800052158. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

