CATS: Cas9-assisted tag switching. A high-throughput method for exchanging genomic peptide tags in yeast
- PMID: 32156257
- PMCID: PMC7063721
- DOI: 10.1186/s12864-020-6634-9
CATS: Cas9-assisted tag switching. A high-throughput method for exchanging genomic peptide tags in yeast
Abstract
Background: The creation of arrays of yeast strains each encoding a different protein with constant tags is a powerful method for understanding how genes and their proteins control cell function. As genetic tools become more sophisticated there is a need to create custom libraries encoding proteins fused with specialised tags to query gene function. These include protein tags that enable a multitude of added functionality, such as conditional degradation, fluorescent labelling, relocalization or activation and also DNA and RNA tags that enable barcoding of genes or their mRNA products. Tools for making new libraries or modifying existing ones are becoming available, but are often limited by the number of strains they can be realistically applied to or by the need for a particular starting library.
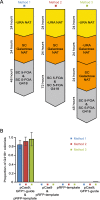
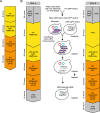
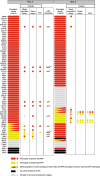
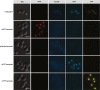
Results: We present a new recombination-based method, CATS - Cas9-Assisted Tag Switching, that switches tags in any existing library of yeast strains. This method employs the reprogrammable RNA guided nuclease, Cas9, to both introduce endogenous double strand breaks into the genome as well as liberating a linear DNA template molecule from a plasmid. It exploits the relatively high efficiency of homologous recombination in budding yeast compared with non-homologous end joining.
Conclusions: The method takes less than 2 weeks, is cost effective and can simultaneously introduce multiple genetic changes, thus providing a rapid, genome-wide approach to genetic modification.
Keywords: Array; CRISPR-Cas9; GFP collection; SPA; Tag switching; Yeast.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Simple CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae.Curr Protoc Mol Biol. 2019 Dec;129(1):e110. doi: 10.1002/cpmb.110. Curr Protoc Mol Biol. 2019. PMID: 31763795 Free PMC article.
-
Expanding the CRISPR/Cas9 Toolbox for Gene Engineering in S. cerevisiae.Curr Microbiol. 2020 Mar;77(3):468-478. doi: 10.1007/s00284-019-01851-0. Epub 2020 Jan 4. Curr Microbiol. 2020. PMID: 31901956
-
A new inducible CRISPR-Cas9 system useful for genome editing and study of double-strand break repair in Candida glabrata.Yeast. 2019 Dec;36(12):723-731. doi: 10.1002/yea.3440. Epub 2019 Sep 5. Yeast. 2019. PMID: 31423617
-
Precision genome editing in the CRISPR era.Biochem Cell Biol. 2017 Apr;95(2):187-201. doi: 10.1139/bcb-2016-0137. Epub 2016 Sep 29. Biochem Cell Biol. 2017. PMID: 28177771 Review.
-
Development and expansion of the CRISPR/Cas9 toolboxes for powerful genome engineering in yeast.Enzyme Microb Technol. 2022 Sep;159:110056. doi: 10.1016/j.enzmictec.2022.110056. Epub 2022 May 7. Enzyme Microb Technol. 2022. PMID: 35561628 Review.
Cited by
-
Forced association of SARS-CoV-2 proteins with the yeast proteome perturb vesicle trafficking.Microb Cell. 2021 Oct 27;8(12):280-296. doi: 10.15698/mic2021.12.766. eCollection 2021 Dec 6. Microb Cell. 2021. PMID: 34909432 Free PMC article.
-
Assembly and delivery of large DNA via chromosome elimination in yeast.Nat Protoc. 2025 Jul 10. doi: 10.1038/s41596-025-01214-z. Online ahead of print. Nat Protoc. 2025. PMID: 40640356 Review.
-
CRI-SPA: a high-throughput method for systematic genetic editing of yeast libraries.Nucleic Acids Res. 2023 Sep 22;51(17):e91. doi: 10.1093/nar/gkad656. Nucleic Acids Res. 2023. PMID: 37572348 Free PMC article.
References
-
- Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H. Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2015;4:585–594. - PubMed
-
- De Antoni A, Gallwitz D. A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene. 2000;246:179–185. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

