A Sustained Immune Response Supports Long-Term Antiviral Immune Priming in the Pacific Oyster, Crassostrea gigas
- PMID: 32156821
- PMCID: PMC7064767
- DOI: 10.1128/mBio.02777-19
A Sustained Immune Response Supports Long-Term Antiviral Immune Priming in the Pacific Oyster, Crassostrea gigas
Abstract
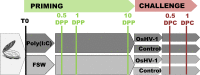
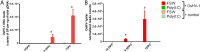
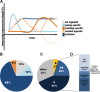
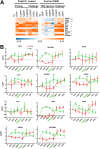
Over the last decade, innate immune priming has been evidenced in many invertebrate phyla. If mechanistic models have been proposed, molecular studies aiming to substantiate these models have remained scarce. We reveal here the transcriptional signature associated with immune priming in the oyster Crassostrea gigas Oysters were fully protected against Ostreid herpesvirus 1 (OsHV-1), a major oyster pathogen, after priming with poly(I·C), which mimics viral double-stranded RNA. Global analysis through RNA sequencing of oyster and viral genes after immune priming and viral infection revealed that poly(I·C) induces a strong antiviral response that impairs OsHV-1 replication. Protection is based on a sustained upregulation of immune genes, notably genes involved in the interferon pathway and apoptosis, which control subsequent viral infection. This persistent antiviral alert state remains active over 4 months and supports antiviral protection in the long term. This acquired resistance mechanism reinforces the molecular foundations of the sustained response model of immune priming. It further opens the way to applications (pseudovaccination) to cope with a recurrent disease that causes dramatic economic losses in the shellfish farming industry worldwide.IMPORTANCE In the last decade, important discoveries have shown that resistance to reinfection can be achieved without a functional adaptive immune system, introducing the concept of innate immune memory in invertebrates. However, this field has been constrained by the limited number of molecular mechanisms evidenced to support these phenomena. Taking advantage of an invertebrate species, the Pacific oyster (Crassostrea gigas), in which we evidenced one of the longest and most effective periods of protection against viral infection observed in an invertebrate, we provide the first comprehensive transcriptomic analysis of antiviral innate immune priming. We show that priming with poly(I·C) induced a massive upregulation of immune-related genes, which control subsequent viral infection, and it was maintained for over 4 months after priming. This acquired resistant mechanism reinforces the molecular foundations of the sustained response model of immune priming. It opens the way to pseudovaccination to prevent the recurrent diseases that currently afflict economically or ecologically important invertebrates.
Keywords: OsHV-1; POMS; antiviral response; immune memory; innate immunity; interferon; oyster; poly(I·C); priming; transcriptomic.
Copyright © 2020 Lafont et al.
Figures



Comment in
-
Towards a Mechanism for Poly(I·C) Antiviral Priming in Oysters.mBio. 2020 Mar 24;11(2):e00407-20. doi: 10.1128/mBio.00407-20. mBio. 2020. PMID: 32209685 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

