Decorating bacteria with self-assembled synthetic receptors
- PMID: 32157077
- PMCID: PMC7064574
- DOI: 10.1038/s41467-020-14336-7
Decorating bacteria with self-assembled synthetic receptors
Abstract
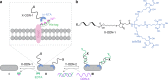
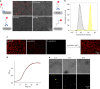
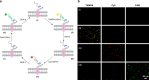
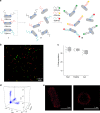
The responses of cells to their surroundings are mediated by the binding of cell surface proteins (CSPs) to extracellular signals. Such processes are regulated via dynamic changes in the structure, composition, and expression levels of CSPs. In this study, we demonstrate the possibility of decorating bacteria with artificial, self-assembled receptors that imitate the dynamic features of CSPs. We show that the local concentration of these receptors on the bacterial membrane and their structure can be reversibly controlled using suitable chemical signals, in a way that resembles changes that occur with CSP expression levels or posttranslational modifications (PTMs), respectively. We also show that these modifications can endow the bacteria with programmable properties, akin to the way CSP responses can induce cellular functions. By programming the bacteria to glow, adhere to surfaces, or interact with proteins or mammalian cells, we demonstrate the potential to tailor such biomimetic systems for specific applications.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Development of a synthetic receptor protein for sensing inflammatory mediators interferon-γ and tumor necrosis factor-α.Biotechnol Bioeng. 2016 Mar;113(3):492-500. doi: 10.1002/bit.25832. Epub 2016 Jan 15. Biotechnol Bioeng. 2016. PMID: 26370067
-
Escherichia coli ZipA Organizes FtsZ Polymers into Dynamic Ring-Like Protofilament Structures.mBio. 2018 Jun 19;9(3):e01008-18. doi: 10.1128/mBio.01008-18. mBio. 2018. PMID: 29921670 Free PMC article.
-
Insights into the Phylogeny and Evolution of Cold Shock Proteins: From Enteropathogenic Yersinia and Escherichia coli to Eubacteria.Int J Mol Sci. 2019 Aug 20;20(16):4059. doi: 10.3390/ijms20164059. Int J Mol Sci. 2019. PMID: 31434224 Free PMC article.
-
Membrane assembly in bacteria.Subcell Biochem. 1994;22:327-59. doi: 10.1007/978-1-4615-2401-4_10. Subcell Biochem. 1994. PMID: 8146886 Review. No abstract available.
-
Advances in understanding bacterial outer-membrane biogenesis.Nat Rev Microbiol. 2006 Jan;4(1):57-66. doi: 10.1038/nrmicro1322. Nat Rev Microbiol. 2006. PMID: 16357861 Review.
Cited by
-
Chemical Artificial Internalizing Receptors for Primary T Cells.Adv Sci (Weinh). 2020 Jul 26;7(18):2001395. doi: 10.1002/advs.202001395. eCollection 2020 Sep. Adv Sci (Weinh). 2020. PMID: 32999846 Free PMC article.
-
Synthetic Artificial Apoptosis-Inducing Receptor for On-Demand Deactivation of Engineered Cells.Adv Sci (Weinh). 2021 May 1;8(13):2004432. doi: 10.1002/advs.202004432. eCollection 2021 Jul. Adv Sci (Weinh). 2021. PMID: 36246165 Free PMC article.
-
Chemically programmable bacterial probes for the recognition of cell surface proteins.Mater Today Bio. 2023 May 23;20:100669. doi: 10.1016/j.mtbio.2023.100669. eCollection 2023 Jun. Mater Today Bio. 2023. PMID: 37334185 Free PMC article.
-
Encrypting messages with artificial bacterial receptors.Beilstein J Org Chem. 2020 Nov 12;16:2749-2756. doi: 10.3762/bjoc.16.225. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 33224301 Free PMC article.
-
A molecular toolkit for heterologous protein secretion across Bacteroides species.Nat Commun. 2024 Nov 11;15(1):9741. doi: 10.1038/s41467-024-53845-7. Nat Commun. 2024. PMID: 39528443 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

