Using the circulating proteome to assess type I interferon activity in systemic lupus erythematosus
- PMID: 32157125
- PMCID: PMC7064569
- DOI: 10.1038/s41598-020-60563-9
Using the circulating proteome to assess type I interferon activity in systemic lupus erythematosus
Abstract
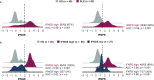
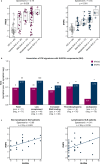
Type I interferon (IFN) drives pathology in systemic lupus erythematosus (SLE) and can be tracked via IFN-inducible transcripts in blood. Here, we examined whether measurement of circulating proteins, which enter the bloodstream from inflamed tissues, also offers insight into global IFN activity. Using a novel protocol we generated 1,132 aptamer-based protein measurements from anti-dsDNApos SLE blood samples and derived an IFN protein signature (IFNPS) that approximates the IFN 21-gene signature (IFNGS). Of 82 patients with SLE, IFNPS was elevated for 89% of IFNGS-high patients (49/55) and 26% of IFNGS-low patients (7/27). IFNGS-high/IFNPS-high patients exhibited activated NK, CD4, and CD8 T cells, while IFNPS-high only patients did not. IFNPS correlated with global disease activity in lymphopenic and non-lymphopenic patients and decreased following type I IFN neutralisation with anifrolumab in the SLE phase IIb study, MUSE. In summary, we developed a protein signature that reflects IFNGS and identifies a new subset of patients with SLE who have IFN activity.
Conflict of interest statement
M.A.Sm. and W.A.R. were employees of AstraZeneca at the time that this analysis was conducted, hold stock/shares in AstraZeneca PLC, and are currently employees of Viela Bio. S.H. was supported by the NIH/NIAMS during the conduct of the study and supported [in part] by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. C.C.C., K.Z., S.R., W.I.W., D.S., and K.S. are employees of AstraZeneca and hold stock/shares in AstraZeneca PLC. M.A.Sa. was an employee of AstraZeneca at the time that this analysis was conducted, holds stock/shares in AstraZeneca PLC, and is currently an employee of Bristol-Myers Squibb. M.J.K. was supported by the NIH/NIAMS during the conduct of the study. K.A.C. was an employee of AstraZeneca at the time that this analysis was conducted and is currently an employee of Bristol-Myers Squibb. R.S. reports intramural funding from National Institutes of Health (NIH) during the conduct of the study; and has been at Novartis Institutes of BioMedical Research since June 2018. A.S., L.G.R., F.W.M., and Z.M. have nothing to declare.
Figures





References
-
- Higgs BW, et al. A phase 1b clinical trial evaluating sifalimumab, an anti-IFN-alpha monoclonal antibody, shows target neutralisation of a type I IFN signature in blood of dermatomyositis and polymyositis patients. Ann. Rheum. Dis. 2014;73:256–262. doi: 10.1136/annrheumdis-2012-202794. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

