Eosinophil accumulation in postnatal lung is specific to the primary septation phase of development
- PMID: 32157178
- PMCID: PMC7064572
- DOI: 10.1038/s41598-020-61420-5
Eosinophil accumulation in postnatal lung is specific to the primary septation phase of development
Abstract
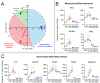
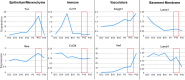
Type 2 immune cells and eosinophils are transiently present in the lung tissue not only in pathology (allergic disease, parasite expulsion) but also during normal postnatal development. However, the lung developmental processes underlying airway recruitment of eosinophils after birth remain unexplored. We determined that in mice, mature eosinophils are transiently recruited to the lung during postnatal days 3-14, which specifically corresponds to the primary septation/alveolarization phase of lung development. Developmental eosinophils peaked during P10-14 and exhibited Siglec-Fmed/highCD11c-/low phenotypes, similar to allergic asthma models. By interrogating the lung transcriptome and proteome during peak eosinophil recruitment in postnatal development, we identified markers that functionally capture the establishment of the mesenchymal-epithelial interface (Nes, Smo, Wnt5a, Nog) and the deposition of the provisional extracellular matrix (ECM) (Tnc, Postn, Spon2, Thbs2) as a key lung morphogenetic event associating with eosinophils. Tenascin-C (TNC) was identified as one of the key ECM markers in the lung epithelial-mesenchymal interface both at the RNA and protein levels, consistently associating with eosinophils in development and disease in mice and humans. As determined by RNA-seq analysis, naïve murine eosinophils cultured with ECM enriched in TNC significantly induced expression of Siglec-F, CD11c, eosinophil peroxidase, and other markers typical for activated eosinophils in development and allergic inflammatory responses. TNC knockout mice had an altered eosinophil recruitment profile in development. Collectively, our results indicate that lung morphogenetic processes associated with heightened Type 2 immunity are not merely a tissue "background" but specifically guide immune cells both in development and pathology.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

