From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes
- PMID: 32159237
- PMCID: PMC7228358
- DOI: 10.1002/jmv.25754
From SARS and MERS CoVs to SARS-CoV-2: Moving toward more biased codon usage in viral structural and nonstructural genes
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an emerging disease with fatal outcomes. In this study, a fundamental knowledge gap question is to be resolved by evaluating the differences in biological and pathogenic aspects of SARS-CoV-2 and the changes in SARS-CoV-2 in comparison with the two prior major COV epidemics, SARS and Middle East respiratory syndrome (MERS) coronaviruses.
Methods: The genome composition, nucleotide analysis, codon usage indices, relative synonymous codons usage, and effective number of codons (ENc) were analyzed in the four structural genes; Spike (S), Envelope (E), membrane (M), and Nucleocapsid (N) genes, and two of the most important nonstructural genes comprising RNA-dependent RNA polymerase and main protease (Mpro) of SARS-CoV-2, Beta-CoV from pangolins, bat SARS, MERS, and SARS CoVs.
Results: SARS-CoV-2 prefers pyrimidine rich codons to purines. Most high-frequency codons were ending with A or T, while the low frequency and rare codons were ending with G or C. SARS-CoV-2 structural proteins showed 5 to 20 lower ENc values, compared with SARS, bat SARS, and MERS CoVs. This implies higher codon bias and higher gene expression efficiency of SARS-CoV-2 structural proteins. SARS-CoV-2 encoded the highest number of over-biased and negatively biased codons. Pangolin Beta-CoV showed little differences with SARS-CoV-2 ENc values, compared with SARS, bat SARS, and MERS CoV.
Conclusion: Extreme bias and lower ENc values of SARS-CoV-2, especially in Spike, Envelope, and Mpro genes, are suggestive for higher gene expression efficiency, compared with SARS, bat SARS, and MERS CoVs.
Keywords: COVID-19; MERS CoV; SARS-CoV-2; codon bias; nonstructural protein; preferred codons.
© 2020 Wiley Periodicals, Inc.
Figures
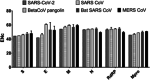
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

