Large-scale in vitro functional testing and novel variant scoring via protein modeling provide insights into alkaline phosphatase activity in hypophosphatasia
- PMID: 32160374
- PMCID: PMC7317754
- DOI: 10.1002/humu.24010
Large-scale in vitro functional testing and novel variant scoring via protein modeling provide insights into alkaline phosphatase activity in hypophosphatasia
Abstract
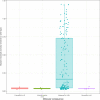
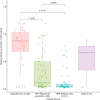
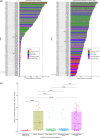
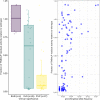
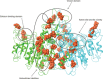
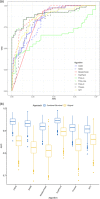
Hypophosphatasia (HPP) is a rare metabolic disorder characterized by low tissue-nonspecific alkaline phosphatase (TNSALP) typically caused by ALPL gene mutations. HPP is heterogeneous, with clinical presentation correlating with residual TNSALP activity and/or dominant-negative effects (DNE). We measured residual activity and DNE for 155 ALPL variants by transient transfection and TNSALP enzymatic activity measurement. Ninety variants showed low residual activity and 24 showed DNE. These results encompass all missense variants with carrier frequencies above 1/25,000 from the Genome Aggregation Database. We used resulting data as a reference to develop a new computational algorithm that scores ALPL missense variants and predicts high/low TNSALP enzymatic activity. Our approach measures the effects of amino acid changes on TNSALP dimer stability with a physics-based implicit solvent energy model. We predict mutation deleteriousness with high specificity, achieving a true-positive rate of 0.63 with false-positive rate of 0, with an area under receiver operating curve (AUC) of 0.9, better than all in silico predictors tested. Combining this algorithm with other in silico approaches can further increase performance, reaching an AUC of 0.94. This study expands our understanding of HPP heterogeneity and genotype/phenotype relationships with the aim of improving clinical ALPL variant interpretation.
Keywords: algorithms; genetic data bases; hypophosphatasia; rare disease; tissue-nonspecific alkaline phosphatase; variant effect prediction.
© 2020 The Authors. Human Mutation published by Wiley Periodicals, Inc.
Conflict of interest statement
G. d. A. and J. R. are employees of Alexion Pharmaceuticals, Inc., the study sponsor, and may own stock and/or stock options in the company. E. M. has received honoraria from Alexion Pharmaceuticals, Inc. C. N. is a former employee and T. S. is a current employee of Schrödinger and they may own stock and/or stock options in the company.
Figures

References
-
- Baumgartner‐Sigl, S. , Haberlandt, E. , Mumm, S. , Scholl‐Burgi, S. , Sergi, C. , Ryan, L. , … Hogler, W. (2007). Pyridoxine‐responsive seizures as the first symptom of infantile hypophosphatasia caused by two novel missense mutations (c.677T>C, p.M226T; c.1112C>T, p.T371I) of the tissue‐nonspecific alkaline phosphatase gene. Bone, 40(6), 1655–1661. 10.1016/j.bone.2007.01.020 - DOI - PubMed
-
- Brun‐Heath, I. , Lia‐Baldini, A. S. , Maillard, S. , Taillandier, A. , Utsch, B. , Nunes, M. E. , … Mornet, E. (2007). Delayed transport of tissue‐nonspecific alkaline phosphatase with missense mutations causing hypophosphatasia. European Journal of Medical Genetics, 50(5), 367–378. 10.1016/j.ejmg.2007.06.005 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

