MYC Dysregulates Mitosis, Revealing Cancer Vulnerabilities
- PMID: 32160543
- PMCID: PMC7085414
- DOI: 10.1016/j.celrep.2020.02.041
MYC Dysregulates Mitosis, Revealing Cancer Vulnerabilities
Abstract
Tumors that overexpress the MYC oncogene are frequently aneuploid, a state associated with highly aggressive cancers and tumor evolution. However, how MYC causes aneuploidy is not well understood. Here, we show that MYC overexpression induces mitotic spindle assembly defects and chromosomal instability (CIN) through effects on microtubule nucleation and organization. Attenuating MYC expression reverses mitotic defects, even in established tumor cell lines, indicating an ongoing role for MYC in CIN. MYC reprograms mitotic gene expression, and we identify TPX2 to be permissive for spindle assembly in MYC-high cells. TPX2 depletion blocks mitotic progression, induces cell death, and prevents tumor growth. Further elevating TPX2 expression reduces mitotic defects in MYC-high cells. MYC and TPX2 expression may be useful biomarkers to stratify patients for anti-mitotic therapies. Our studies implicate MYC as a regulator of mitosis and suggest that blocking MYC activity can attenuate the emergence of CIN and tumor evolution.
Keywords: CIN; MYC; TNBC; TPX2; chromosomal instability; microtubules; mitosis; mitotic spindle assembly; receptor triple-negative breast cancer; synthetic-lethality.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures


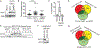
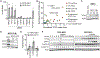



References
-
- Aguirre-Portolés C, Bird AW, Hyman A, Cañamero M, Pérez de Castro I, and Malumbres M (2012). Tpx2 controls spindle integrity, genome stability, and tumor development. Cancer Res. 72, 1518–1528. - PubMed
-
- Amati B, Alevizopoulos K, and Vlach J (1998). Myc and the cell cycle. Front. Biosci 3, d250–d268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

