Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi
- PMID: 32161129
- PMCID: PMC7104180
- DOI: 10.1073/pnas.1919726117
Natural diversity in the predatory behavior facilitates the establishment of a robust model strain for nematode-trapping fungi
Abstract
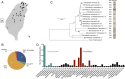
Nematode-trapping fungi (NTF) are a group of specialized microbial predators that consume nematodes when food sources are limited. Predation is initiated when conserved nematode ascaroside pheromones are sensed, followed by the development of complex trapping devices. To gain insights into the coevolution of this interkingdom predator-prey relationship, we investigated natural populations of nematodes and NTF that we found to be ubiquitous in soils. Arthrobotrys species were sympatric with various nematode species and behaved as generalist predators. The ability to sense prey among wild isolates of Arthrobotrys oligospora varied greatly, as determined by the number of traps after exposure to Caenorhabditis elegans While some strains were highly sensitive to C. elegans and the nematode pheromone ascarosides, others responded only weakly. Furthermore, strains that were highly sensitive to the nematode prey also developed traps faster. The polymorphic nature of trap formation correlated with competency in prey killing, as well as with the phylogeny of A. oligospora natural strains, calculated after assembly and annotation of the genomes of 20 isolates. A chromosome-level genome assembly and annotation were established for one of the most sensitive wild isolates, and deletion of the only G-protein β-subunit-encoding gene of A. oligospora nearly abolished trap formation. In summary, our study establishes a highly responsive A. oligospora wild isolate as a model strain for the study of fungus-nematode interactions and demonstrates that trap formation is a fitness character in generalist predators of the nematode-trapping fungus family.
Keywords: G-protein signaling; natural population; nematode-trapping fungi; predator–prey interaction; trap morphogenesis.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Bardgett R. D., van der Putten W. H., Belowground biodiversity and ecosystem functioning. Nature 515, 505–511 (2014). - PubMed
-
- van den Hoogen J., et al. , Soil nematode abundance and functional group composition at a global scale. Nature 572, 194–198 (2019). - PubMed
-
- Vidal-Diez de Ulzurrun G., Hsueh Y. P., Predator-prey interactions of nematode-trapping fungi and nematodes: Both sides of the coin. Appl. Microbiol. Biotechnol. 102, 3939–3949 (2018). - PubMed

