Integrative study of the upper and lower airway microbiome and transcriptome in asthma
- PMID: 32161195
- PMCID: PMC7141394
- DOI: 10.1172/jci.insight.133707
Integrative study of the upper and lower airway microbiome and transcriptome in asthma
Abstract
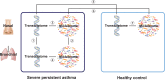
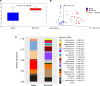
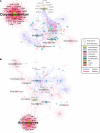
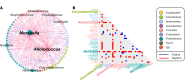
Relatively little is known about interactions between the airway microbiome and airway host transcriptome in asthma. Since asthma affects and is affected by the entire airway, studying the upper (e.g., nasal) and lower (e.g., bronchial) airways together represents a powerful approach to understanding asthma. Here, we performed a systematic, integrative study of the nasal and bronchial microbiomes and nasal and bronchial host transcriptomes of children with severe persistent asthma and healthy controls. We found that (a) the microbiomes and host transcriptomes of asthmatic children are each distinct by site (nasal versus bronchial); (b) among asthmatic children, Moraxella and Alloiococcus are hub genera in the nasal microbiome, while there are no hubs among bronchial genera; (c) bronchial Actinomyces is negatively associated with bronchial genes for inflammation, suggesting Actinomyces may be protective; (d) compared with healthy children, asthmatic children express more nasal genes for ciliary function and harbor more nasal Streptococcus; and (e) nasal genera such as Corynebacterium are negatively associated with significantly more nasal genes for inflammation in healthy versus asthmatic children, suggesting a potentially stronger protective role for such nasal genera in healthy versus asthmatic children. Our systematic, integrative study provides a window into host-microbiome associations in asthma.
Keywords: Asthma; Bacterial infections; Microbiology; Pulmonology; Transcription.
Conflict of interest statement
Figures




References
-
- Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386(9995):743–800. doi: 10.1016/S0140-6736(15)60692-4. - DOI - PMC - PubMed

