Emergence of norovirus strains: A tale of two genes
- PMID: 32161666
- PMCID: PMC6875644
- DOI: 10.1093/ve/vez048
Emergence of norovirus strains: A tale of two genes
Abstract
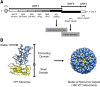
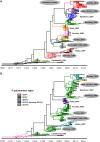
Noroviruses are a very diverse group of viruses that infect different mammalian species. In humans, norovirus is a major cause of acute gastroenteritis. Multiple norovirus infections can occur in a lifetime as the result of limited duration of acquired immunity and cross-protection among different strains. A combination of advances in sequencing methods and improvements on surveillance has provided new insights into norovirus diversification and emergence. The generation of diverse norovirus strains has been associated with (1) point mutations on two different genes: ORF1, encoding the non-structural proteins, and ORF2, encoding the major capsid protein (VP1); and (2) recombination events that create chimeric viruses. While both mechanisms are exploited by all norovirus strains, individual genotypes utilize each mechanism differently to emerge and persist in the human population. GII.4 noroviruses (the most prevalent genotype in humans) present an accumulation of amino acid mutations on VP1 resulting in the chronological emergence of new variants. In contrast, non-GII.4 noroviruses present co-circulation of different variants over long periods with limited changes on their VP1. Notably, genetic diversity of non-GII.4 noroviruses is mostly related to the high number of recombinant strains detected in humans. While it is difficult to determine the precise mechanism of emergence of epidemic noroviruses, observations point to multiple factors that include host-virus interactions and changes on two regions of the genome (ORF1 and ORF2). Larger datasets of viral genomes are needed to facilitate comparison of epidemic strains and those circulating at low levels in the population. This will provide a better understanding of the mechanism of norovirus emergence and persistence.
Keywords: caliciviruses; evolution; gastroenteritis; norovirus; recombination.
Published by Oxford University Press 2019. This work is written by a US Government employee and is in the public domain in the US.
Figures


References
-
- Ao Y. et al. (2018) ‘Genetic Analysis of Reemerging GII.P16-GII.2 Noroviruses in 2016–2017 in China’, The Journal of Infectious Diseases, 218: 133–43. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

