Applications of the CRISPR/Cas system beyond gene editing
- PMID: 32161796
- PMCID: PMC6994046
- DOI: 10.1093/biomethods/bpy002
Applications of the CRISPR/Cas system beyond gene editing
Abstract
Since the discovery of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated system (Cas) as a tool for gene editing a plethora of locus-specific as well as genome-wide approaches have been developed that allow efficient and reproducible manipulation of genomic sequences. However, the seemingly unbound potential of CRISPR/Cas does not stop with its utilization as a site-directed nuclease. Mutations in its catalytic centers render Cas9 (dCas9) a universal recruitment platform that can be utilized to control transcription, visualize DNA sequences, investigate in situ proteome compositions and manipulate epigenetic modifications at user-defined genomic loci. In this review, we give a comprehensive introduction and overview of the development, improvement and application of recent dCas9-based approaches.
Keywords: CASPEX; CRISPR imaging; CRISPR/Cas; CRISPRa; CRISPRi; CasID; dCas9.
© The Author(s) 2018. Published by Oxford University Press.
Figures
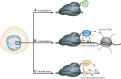
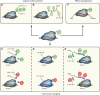

Similar articles
-
Targeted Modulation of Chicken Genes In Vitro Using CRISPRa and CRISPRi Toolkit.Genes (Basel). 2023 Apr 13;14(4):906. doi: 10.3390/genes14040906. Genes (Basel). 2023. PMID: 37107664 Free PMC article.
-
Applications of CRISPR/Cas System to Bacterial Metabolic Engineering.Int J Mol Sci. 2018 Apr 5;19(4):1089. doi: 10.3390/ijms19041089. Int J Mol Sci. 2018. PMID: 29621180 Free PMC article. Review.
-
CRISPR technologies for stem cell engineering and regenerative medicine.Biotechnol Adv. 2019 Dec;37(8):107447. doi: 10.1016/j.biotechadv.2019.107447. Epub 2019 Sep 9. Biotechnol Adv. 2019. PMID: 31513841 Review.
-
Beyond Native Cas9: Manipulating Genomic Information and Function.Trends Biotechnol. 2017 Oct;35(10):983-996. doi: 10.1016/j.tibtech.2017.06.004. Epub 2017 Jul 21. Trends Biotechnol. 2017. PMID: 28739220 Review.
-
Recent Progress and Future Prospect of CRISPR/Cas-Derived Transcription Activation (CRISPRa) System in Plants.Cells. 2022 Sep 28;11(19):3045. doi: 10.3390/cells11193045. Cells. 2022. PMID: 36231007 Free PMC article. Review.
Cited by
-
WeReview: CRISPR Tools-Live Repository of Computational Tools for Assisting CRISPR/Cas Experiments.Bioengineering (Basel). 2019 Jul 25;6(3):63. doi: 10.3390/bioengineering6030063. Bioengineering (Basel). 2019. PMID: 31349743 Free PMC article.
-
CRISPRi for specific inhibition of miRNA clusters and miRNAs with high sequence homology.Sci Rep. 2022 Apr 15;12(1):6297. doi: 10.1038/s41598-022-10336-3. Sci Rep. 2022. PMID: 35428787 Free PMC article.
-
Applications of CRISPR-Cas9 in Alzheimer's Disease and Related Disorders.Int J Mol Sci. 2022 Aug 5;23(15):8714. doi: 10.3390/ijms23158714. Int J Mol Sci. 2022. PMID: 35955847 Free PMC article. Review.
-
Integration of Organoids With CRISPR Screens: A Narrative Review.Biol Cell. 2025 Apr;117(4):e70006. doi: 10.1111/boc.70006. Biol Cell. 2025. PMID: 40223602 Free PMC article. Review.
-
A Leak-Free Inducible CRISPRi/a System for Gene Functional Studies in Plasmodium falciparum.Microbiol Spectr. 2022 Jun 29;10(3):e0278221. doi: 10.1128/spectrum.02782-21. Epub 2022 May 5. Microbiol Spectr. 2022. PMID: 35510853 Free PMC article.
References
-
- Samson JE, Magadan AH, Sabri M. et al. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol 2013;11:675–87. - PubMed
-
- Barrangou R, Fremaux C, Deveau H. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007;315:1709.. - PubMed
-
- Wiedenheft B, Sternberg SH, Doudna JA.. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012;482:331–338. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
