Genome-wide DNA hypomethylation shapes nematode pattern-triggered immunity in plants
- PMID: 32162327
- PMCID: PMC7317725
- DOI: 10.1111/nph.16532
Genome-wide DNA hypomethylation shapes nematode pattern-triggered immunity in plants
Abstract
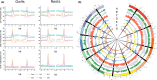
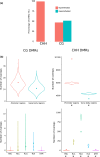
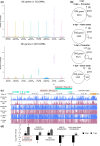
A role for DNA hypomethylation has recently been suggested in the interaction between bacteria and plants; it is unclear whether this phenomenon reflects a conserved response. Treatment of plants of monocot rice and dicot tomato with nematode-associated molecular patterns from different nematode species or bacterial pathogen-associated molecular pattern flg22 revealed global DNA hypomethylation. A similar hypomethylation response was observed during early gall induction by Meloidogyne graminicola in rice. Evidence for the causal impact of hypomethylation on immunity was revealed by a significantly reduced plant susceptibility upon treatment with DNA methylation inhibitor 5-azacytidine. Whole-genome bisulphite sequencing of young galls revealed massive hypomethylation in the CHH context, while not for CG or CHG nucleotide contexts. Further, CHH hypomethylated regions were predominantly associated with gene promoter regions, which was not correlated with activated gene expression at the same time point but, rather, was correlated with a delayed transcriptional gene activation. Finally, the relevance of CHH hypomethylation in plant defence was confirmed in rice mutants of the RNA-directed DNA methylation pathway and DECREASED DNA METHYLATION 1. We demonstrated that DNA hypomethylation is associated with reduced susceptibility in rice towards root-parasitic nematodes and is likely to be part of the basal pattern-triggered immunity response in plants.
Keywords: Meloidogyne graminicola; Oryza sativa; DNA hypomethylation; RdDM; basal defence; nematodes; pattern-triggered immunity; rice.
© 2020 The Authors. New Phytologist © 2020 New Phytologist Trust.
Figures



References
-
- Bouvet GF, Jacobi V, Plourde KV, Bernier L. 2008. Stress‐induced mobility of OPHIO1 and OPHIO2, DNA transposons of the Dutch elm disease fungi. Fungal Genetics and Biology 45: 565–578. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

