Resolving Binding Events on the Multifunctional Human Serum Albumin
- PMID: 32162429
- PMCID: PMC7318646
- DOI: 10.1002/cmdc.202000069
Resolving Binding Events on the Multifunctional Human Serum Albumin
Abstract
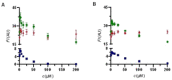
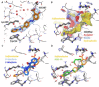
Physiological processes rely on initial recognition events between cellular components and other molecules or modalities. Biomolecules can have multiple sites or mode of interaction with other molecular entities, so that a resolution of the individual binding events in terms of spatial localization as well as association and dissociation kinetics is required for a meaningful description. Here we describe a trichromatic fluorescent binding- and displacement assay for simultaneous monitoring of three individual binding sites in the important transporter and binding protein human serum albumin. Independent investigations of binding events by X-ray crystallography and time-resolved dynamics measurements (switchSENSE technology) confirm the validity of the assay, the localization of binding sites and furthermore reveal conformational changes associated with ligand binding. The described assay system allows for the detailed characterization of albumin-binding drugs and is therefore well-suited for prediction of drug-drug and drug-food interactions. Moreover, conformational changes, usually associated with binding events, can also be analyzed.
Keywords: albumin binding; drug interactions; kinetics investigations; multicolor assays; switchSENSE technology.
© 2020 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

