Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer
- PMID: 32162812
- PMCID: PMC7066697
- DOI: 10.1634/theoncologist.2019-0441
Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer
Abstract
Background: Direct comparisons between Guardant360 (G360) circulating tumor DNA (ctDNA) and FoundationOne (F1) tumor biopsy genomic profiling in metastatic colorectal cancer (mCRC) are limited. We aim to assess the concordance across overlapping genes tested in both F1 and G360 in patients with mCRC.
Materials and methods: We retrospectively analyzed 75 patients with mCRC who underwent G360 and F1 testing. We evaluated the concordance among gene mutations tested by both G360 and F1 among three categories of patients: untreated, treated without, and treated with EGFR inhibitors, while considering the clonal and/or subclonal nature of each genomic alteration.
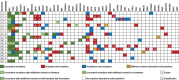
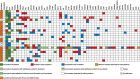
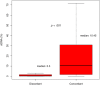
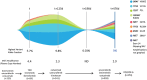
Results: There was a high rate of concordance in APC, TP53, KRAS, NRAS, and BRAF mutations in the treatment-naive and non-anti-EGFR-treated cohorts. There was increased discordance in the anti-EGFR treated patients in three drivers of anti-EGFR resistance: KRAS, NRAS, and EGFR somatic mutations. Based on percentage of ctDNA, discordant somatic mutations were mostly subclonal instead of clonal and may have limited clinical significance. Most discordant amplifications noted on G360 showed the magnitude below the top decile, occurred in all three cohorts of patients, and were of unknown clinical significance. Serial ctDNA in anti-EGFR treated patients showed the emergence of multiple new alterations that affected the EGFR pathway: EGFR and RAS mutations and MET, RAS, and BRAF amplifications.
Conclusion: G360 Next-Generation Sequencing platform may be used as an alternative to F1 to detect targetable somatic alterations in non-anti-EGFR treated mCRC, but larger prospective studies are needed to further validate our findings.
Implications for practice: Genomic analysis of tissue biopsy is currently the optimal method for identifying DNA genomic alterations to help physicians target specific genes but has many disadvantages that may be mitigated by a circulating free tumor DNA (ctDNA) assay. This study showed a high concordance rate in certain gene mutations in patients who were treatment naive and treated with non-anti-EGFR therapy prior to ctDNA testing. This suggests that ctDNA genomic analysis may potentially be used as an alternative to tumor biopsy to identify appropriate patients for treatment selection in mCRC, but larger prospective studies are needed to further validate concordance among tissue and ctDNA tumor profiling.
Keywords: Circulating tumor DNA; Colorectal cancer; Concordance; EGFR; NGS.
© AlphaMed Press 2019.
Conflict of interest statement
Figures

Similar articles
-
Comprehensive genomic profiling by liquid biopsy captures tumor heterogeneity and identifies cancer vulnerabilities in patients with RAS/BRAFV600E wild-type metastatic colorectal cancer in the CAPRI 2-GOIM trial.Ann Oncol. 2024 Dec;35(12):1105-1115. doi: 10.1016/j.annonc.2024.08.2334. Epub 2024 Aug 29. Ann Oncol. 2024. PMID: 39214459
-
Diagnostic Strategies toward Clinical Implementation of Liquid Biopsy RAS/BRAF Circulating Tumor DNA Analyses in Patients with Metastatic Colorectal Cancer.J Mol Diagn. 2020 Dec;22(12):1430-1437. doi: 10.1016/j.jmoldx.2020.09.002. Epub 2020 Sep 19. J Mol Diagn. 2020. PMID: 32961317
-
High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program.Cancer. 2021 Aug 15;127(16):3019-3028. doi: 10.1002/cncr.33571. Epub 2021 Apr 7. Cancer. 2021. PMID: 33826761
-
Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer.Oncologist. 2018 Nov;23(11):1310-1318. doi: 10.1634/theoncologist.2017-0621. Epub 2018 Apr 26. Oncologist. 2018. PMID: 29700206 Free PMC article. Review.
-
Next-Generation Sequencing of Circulating Tumor DNA Can Optimize Second-Line Treatment in RAS Wild-Type Metastatic Colorectal Cancer after Progression on anti-EGFR Therapy: Time to Rethink Our Approach.Oncol Res Treat. 2022;45(4):216-221. doi: 10.1159/000521845. Epub 2022 Jan 9. Oncol Res Treat. 2022. PMID: 34999585 Review.
Cited by
-
Liquid Biopsy for Patient Characterization in Cardiovascular Disease: Verification against Markers of Cytochrome P450 and P-Glycoprotein Activities.Clin Pharmacol Ther. 2022 Jun;111(6):1268-1277. doi: 10.1002/cpt.2576. Epub 2022 Mar 28. Clin Pharmacol Ther. 2022. PMID: 35262906 Free PMC article.
-
Response prediction by mutation- or methylation-specific detection of ctDNA dynamics in pretreated metastatic colorectal cancer.Ther Adv Med Oncol. 2023 Sep 29;15:17588359231200462. doi: 10.1177/17588359231200462. eCollection 2023. Ther Adv Med Oncol. 2023. PMID: 37786537 Free PMC article.
-
Clinical Utility of a Cell-Free DNA Assay in Patients With Colorectal Cancer.Front Oncol. 2021 Mar 19;11:589673. doi: 10.3389/fonc.2021.589673. eCollection 2021. Front Oncol. 2021. PMID: 33816227 Free PMC article.
-
Current Clinical Practice of Precision Medicine Using Comprehensive Genomic Profiling Tests in Biliary Tract Cancer in Japan.Curr Oncol. 2022 Sep 30;29(10):7272-7284. doi: 10.3390/curroncol29100573. Curr Oncol. 2022. PMID: 36290850 Free PMC article. Review.
-
The Pan-Tumor Landscape of Targetable Kinase Fusions in Circulating Tumor DNA.Clin Cancer Res. 2022 Feb 15;28(4):728-737. doi: 10.1158/1078-0432.CCR-21-2136. Clin Cancer Res. 2022. PMID: 34753780 Free PMC article.
References
-
- Burrell RA, McGranahan N, Bartek J et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013;501:338–345. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

