Inherited human IFN-γ deficiency underlies mycobacterial disease
- PMID: 32163377
- PMCID: PMC7260033
- DOI: 10.1172/JCI135460
Inherited human IFN-γ deficiency underlies mycobacterial disease
Abstract
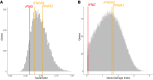
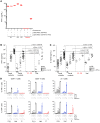
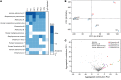
Mendelian susceptibility to mycobacterial disease (MSMD) is characterized by a selective predisposition to clinical disease caused by the Bacille Calmette-Guérin (BCG) vaccine and environmental mycobacteria. The known genetic etiologies of MSMD are inborn errors of IFN-γ immunity due to mutations of 15 genes controlling the production of or response to IFN-γ. Since the first MSMD-causing mutations were reported in 1996, biallelic mutations in the genes encoding IFN-γ receptor 1 (IFN-γR1) and IFN-γR2 have been reported in many patients of diverse ancestries. Surprisingly, mutations of the gene encoding the IFN-γ cytokine itself have not been reported, raising the remote possibility that there might be other agonists of the IFN-γ receptor. We describe 2 Lebanese cousins with MSMD, living in Kuwait, who are both homozygous for a small deletion within the IFNG gene (c.354_357del), causing a frameshift that generates a premature stop codon (p.T119Ifs4*). The mutant allele is loss of expression and loss of function. We also show that the patients' herpesvirus Saimiri-immortalized T lymphocytes did not produce IFN-γ, a phenotype that can be rescued by retrotransduction with WT IFNG cDNA. The blood T and NK lymphocytes from these patients also failed to produce and secrete detectable amounts of IFN-γ. Finally, we show that human IFNG has evolved under stronger negative selection than IFNGR1 or IFNGR2, suggesting that it is less tolerant to heterozygous deleterious mutations than IFNGR1 or IFNGR2. This may account for the rarity of patients with autosomal-recessive, complete IFN-γ deficiency relative to patients with complete IFN-γR1 and IFN-γR2 deficiencies.
Keywords: Genetic diseases; Genetics; Immunology.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

