mRNA Vaccines Encoding the HA Protein of Influenza A H1N1 Virus Delivered by Cationic Lipid Nanoparticles Induce Protective Immune Responses in Mice
- PMID: 32164372
- PMCID: PMC7157730
- DOI: 10.3390/vaccines8010123
mRNA Vaccines Encoding the HA Protein of Influenza A H1N1 Virus Delivered by Cationic Lipid Nanoparticles Induce Protective Immune Responses in Mice
Abstract
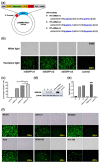
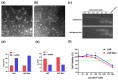
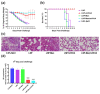
The design of the mRNA vaccine involves the selection of in vitro transcription (IVT) systems and nonviral delivery vectors. This study aimed to verify the effect of 5' and 3' untranslated region (UTR) sequences on the translation efficiency of mRNA. Three modes of IVT-mRNA systems (IVT-mRNA-n1/n2/n3) with diverse UTRs were constructed, and EGFP (enhanced green fluorescent protein) and HA (hemagglutinin) gene of H3N2 influenza virus were introduced into each of them. The results showed that the mode of 5' and 3' UTRs originating from human β-globulin was better than the mode of UTRs from human α-globulin, and the n3 mode was the best. mEGFP-n3, mH3HA-n3, and mLuciferease-n3 were prepared to compare the effect of cationic lipid nanoparticle (LNP) with that of mannose-conjugated LNP (LNP-Man) on the efficiency of gene delivery. The results showed that the effect of LNP-Man was better than that of LNP both in vitro and in vivo. Choosing appropriate ligands might help in vaccine design. After selecting the IVT-mRNA-n3 system and delivery vectors, mRNA vaccines were constructed against the H1N1 influenza virus, and C57BL/6 mice were immunized through intranasal administration. The results showed that mRNA vaccines could elicit both humoral and cellular immune responses and completely protect mice from the tenfold LD50 H1N1 influenza virus challenge.
Keywords: cationic lipid nanoparticles; influenza A H1N1 virus; intranasal administration; mRNA vaccine; mannose.
Conflict of interest statement
The authors declare no conflicts of interest. The sponsors had no role in the design, execution, interpretation, or writing of the study.
Figures

References
-
- Brazzoli M., Magini D., Bonci A., Buccato S., Giovani C., Kratzer R., Zurli V., Mangiavacchi S., Casini D., Brito L.M., et al. Induction of Broad-Based Immunity and Protective Efficacy by Self-amplifying mRNA Vaccines Encoding Influenza Virus Hemagglutinin. J. Virol. 2016;90:332–344. doi: 10.1128/JVI.01786-15. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

