Identification of the regulatory networks and hub genes controlling alfalfa floral pigmentation variation using RNA-sequencing analysis
- PMID: 32164566
- PMCID: PMC7068929
- DOI: 10.1186/s12870-020-2322-9
Identification of the regulatory networks and hub genes controlling alfalfa floral pigmentation variation using RNA-sequencing analysis
Abstract
Background: To understand the gene expression networks controlling flower color formation in alfalfa, flowers anthocyanins were identified using two materials with contrasting flower colors, namely Defu and Zhongtian No. 3, and transcriptome analyses of PacBio full-length sequencing combined with RNA sequencing were performed, across four flower developmental stages.
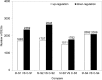
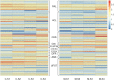
Results: Malvidin and petunidin glycoside derivatives were the major anthocyanins in the flowers of Defu, which were lacking in the flowers of Zhongtian No. 3. The two transcriptomic datasets provided a comprehensive and systems-level view on the dynamic gene expression networks underpinning alfalfa flower color formation. By weighted gene coexpression network analyses, we identified candidate genes and hub genes from the modules closely related to floral developmental stages. PAL, 4CL, CHS, CHR, F3'H, DFR, and UFGT were enriched in the important modules. Additionally, PAL6, PAL9, 4CL18, CHS2, 4 and 8 were identified as hub genes. Thus, a hypothesis explaining the lack of purple color in the flower of Zhongtian No. 3 was proposed.
Conclusions: These analyses identified a large number of potential key regulators controlling flower color pigmentation, thereby providing new insights into the molecular networks underlying alfalfa flower development.
Keywords: Alfalfa; Cream color; Floral pigmentation; Hub gene; PacBio Iso-Seq; Transcriptome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
MeSH terms
Grants and funding
- 31700338/National Natural Science Foundation of China
- 31860118/National Natural Science Foundation of China
- 1610322019012/Fundamental Research Funds for the Central Public-interest Scientific Institution
- 1610322019013/Fundamental Research Funds for the Central Public-interest Scientific Institution
- CAASASTIP-2019-LIHPS-08/Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences
LinkOut - more resources
Full Text Sources
Miscellaneous

