A transcriptional signature accurately identifies Aspergillus Infection across healthy and immunosuppressed states
- PMID: 32165060
- PMCID: PMC7170547
- DOI: 10.1016/j.trsl.2020.02.005
A transcriptional signature accurately identifies Aspergillus Infection across healthy and immunosuppressed states
Abstract
Invasive aspergillosis (IA) is a major cause of critical illness in immunocompromised (IC) patients. However, current fungal tests are limited. Disease-specific gene expression patterns in circulating host cells show promise as novel diagnostics, however it is unknown whether such a 'signature' exists for IA and the effect of iatrogenic immunosuppression on any such biomarkers. Male BALB/c mice were separated into 6 experimental groups based on Aspergillus fumigatus inhalational exposure and IC status (no immunosuppression, cyclophosphamide, and corticosteroids). Mice were sacrificed 4 days postinfection. Whole blood was assayed for transcriptomic responses in peripheral white blood cells via microarray. An elastic net regularized logistic regression was employed to develop classifiers of IA based on gene expression. Aspergillus infection triggers a powerful response in non-IC hosts with 2718 genes differentially expressed between IA and controls. We generated a 146-gene classifier able to discriminate between non-IC infected and uninfected mice with an AUC of 1. However, immunosuppressive medications exhibited a confounding effect on this transcriptomic classifier. After controlling for the genomic effects of immunosuppression, we were able to generate a 187-gene classifier with an AUC of 0.92 in the absence of immunosuppression, 1 with cyclophosphamide, and 0.9 with steroids. The host transcriptomic response to IA is robust and conserved. Pharmacologic perturbation of the host immune response has powerful effects on classifier performance and must be considered when developing such novel diagnostics. When appropriately designed, host-derived peripheral blood transcriptomic responses demonstrate the ability to accurately diagnose Aspergillus infection, even in the presence of immunosuppression.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have read the journal's policy on disclosure of potential conflicts of interest. All authors have disclosed any financial or personal relationship with organizations that could potentially be perceived as influencing the described research. J.M.S., M.T.M., A.K.Z., J.L.M, and D.L.C. are listed on pending patent T-006795.
Figures
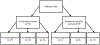

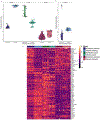

References
-
- De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;46(12):1813–21. - PMC - PubMed
-
- Blot SI, Taccone FS, Van den Abeele AM, et al. A clinical algorithm to diagnose invasive pulmonary aspergillosis in critically ill patients. American journal of respiratory and critical care medicine. 2012;186(1):56–64. - PubMed
-
- Kami M, Murashige N, Fujihara T, Sakagami N, Tanaka Y. The mechanism for low yield of blood culture in invasive aspergillosis; the clinical importance of antigen detection tests revisited. Bone marrow transplantation. 2005;36(1):85–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

