Root transcriptome reveals efficient cell signaling and energy conservation key to aluminum toxicity tolerance in acidic soil adapted rice genotype
- PMID: 32165659
- PMCID: PMC7067865
- DOI: 10.1038/s41598-020-61305-7
Root transcriptome reveals efficient cell signaling and energy conservation key to aluminum toxicity tolerance in acidic soil adapted rice genotype
Abstract
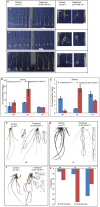
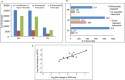
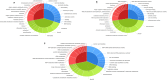
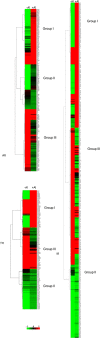
Aluminium (Al) toxicity is the single most important contributing factor constraining crop productivity in acidic soils. Hydroponics based screening of three rice genotypes, a tolerant (ARR09, AR), a susceptible (IR 1552, IR) and an acid soil adapted landrace (Theruvii, TH) revealed that AR accumulates less Al and shows minimum decrease in shoot and root biomass under Al toxicity conditions when compared with IR. Transcriptome data generated on roots (grown in presence or absence of Al) led to identification of ~1500 transcripts per genotype with percentage annotation ranging from 21.94% (AR) to 29.94% (TH). A total of 511, 804 and 912 DEGs were identified in genotypes AR, IR and TH, respectively. IR showed upregulation of transcripts involved in exergonic processes. AR appears to conserve energy by downregulating key genes of glycolysis pathway and maintaining transcript levels of key exergonic step enzymes under Al stress. The tolerance in AR appears to be as a result of novel mechanism as none of the reported Al toxicity genes or QTLs overlap with significant DEGs. Components of signal transduction and regulatory machinery like transcripts encoding zinc finger protein, calcieurin binding protein and cell wall associated transcripts are among the highly upregulated DEGs in AR, suggesting increased and better signal transduction in response to Al stress in tolerant rice. Sequencing of NRAT1 and glycine-rich protein A3 revealed distinct haplotype for indica type AR. The newly identified components of Al tolerance will help in designing molecular breeding tools to enhance rice productivity in acidic soils.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. doi: 10.1007/BF00009558. - DOI
-
- Kumar M, et al. Variable lime requirement based on differences in organic matter content of iso-acidic soils. Indian J. Hill Farming. 2012;25:26–30.
-
- Yumnam JS, Rai M, Tyagi W. Allele mining across two low-P tolerant genes PSTOL1 and PupK20-2 reveals novel haplotypes in rice genotypes adapted to acidic soils. Plant Genet Resour-C. 2017;15:221–229. doi: 10.1017/S1479262115000544. - DOI
-
- Abate E, Hussien S, Laing M, Mengistu F. Aluminium toxicity tolerance in cereals: Mechanisms, genetic control and breeding methods. F. Afr. J. Agric. Res. 2013;8:711–722.
-
- Ma JF, Chen ZC, Shen RF. Molecular mechanisms of Al tolerance in gramineous plants. Plant Soil. 2014;381:1–12. doi: 10.1007/s11104-014-2073-1. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

