Mutation effect estimation on protein-protein interactions using deep contextualized representation learning
- PMID: 32166223
- PMCID: PMC7059401
- DOI: 10.1093/nargab/lqaa015
Mutation effect estimation on protein-protein interactions using deep contextualized representation learning
Abstract
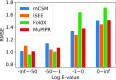
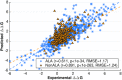
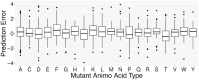
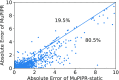
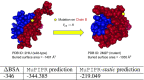
The functional impact of protein mutations is reflected on the alteration of conformation and thermodynamics of protein-protein interactions (PPIs). Quantifying the changes of two interacting proteins upon mutations is commonly carried out by computational approaches. Hence, extensive research efforts have been put to the extraction of energetic or structural features on proteins, followed by statistical learning methods to estimate the effects of mutations on PPI properties. Nonetheless, such features require extensive human labors and expert knowledge to obtain, and have limited abilities to reflect point mutations. We present an end-to-end deep learning framework, MuPIPR (Mutation Effects in Protein-protein Interaction PRediction Using Contextualized Representations), to estimate the effects of mutations on PPIs. MuPIPR incorporates a contextualized representation mechanism of amino acids to propagate the effects of a point mutation to surrounding amino acid representations, therefore amplifying the subtle change in a long protein sequence. On top of that, MuPIPR leverages a Siamese residual recurrent convolutional neural encoder to encode a wild-type protein pair and its mutation pair. Multi-layer perceptron regressors are applied to the protein pair representations to predict the quantifiable changes of PPI properties upon mutations. Experimental evaluations show that, with only sequence information, MuPIPR outperforms various state-of-the-art systems on estimating the changes of binding affinity for SKEMPI v1, and offers comparable performance on SKEMPI v2. Meanwhile, MuPIPR also demonstrates state-of-the-art performance on estimating the changes of buried surface areas. The software implementation is available at https://github.com/guangyu-zhou/MuPIPR.
© The Author(s) 2019. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures



Similar articles
-
Multifaceted protein-protein interaction prediction based on Siamese residual RCNN.Bioinformatics. 2019 Jul 15;35(14):i305-i314. doi: 10.1093/bioinformatics/btz328. Bioinformatics. 2019. PMID: 31510705 Free PMC article.
-
ELECTRA-DTA: a new compound-protein binding affinity prediction model based on the contextualized sequence encoding.J Cheminform. 2022 Mar 15;14(1):14. doi: 10.1186/s13321-022-00591-x. J Cheminform. 2022. PMID: 35292100 Free PMC article.
-
Multimodal deep representation learning for protein interaction identification and protein family classification.BMC Bioinformatics. 2019 Dec 2;20(Suppl 16):531. doi: 10.1186/s12859-019-3084-y. BMC Bioinformatics. 2019. PMID: 31787089 Free PMC article.
-
Convolutional Neural Networks for ATC Classification.Curr Pharm Des. 2018;24(34):4007-4012. doi: 10.2174/1381612824666181112113438. Curr Pharm Des. 2018. PMID: 30417778 Review.
-
Targeting Virus-host Protein Interactions: Feature Extraction and Machine Learning Approaches.Curr Drug Metab. 2019;20(3):177-184. doi: 10.2174/1389200219666180829121038. Curr Drug Metab. 2019. PMID: 30156155 Review.
Cited by
-
Structure-Guided Computational Approaches to Unravel Druggable Proteomic Landscape of Mycobacterium leprae.Front Mol Biosci. 2021 May 7;8:663301. doi: 10.3389/fmolb.2021.663301. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026836 Free PMC article. Review.
-
EpitopeVec: linear epitope prediction using deep protein sequence embeddings.Bioinformatics. 2021 Dec 7;37(23):4517-4525. doi: 10.1093/bioinformatics/btab467. Bioinformatics. 2021. PMID: 34180989 Free PMC article.
-
LambdaPP: Fast and accessible protein-specific phenotype predictions.Protein Sci. 2023 Jan;32(1):e4524. doi: 10.1002/pro.4524. Protein Sci. 2023. PMID: 36454227 Free PMC article.
-
ProstaNet: A Novel Geometric Vector Perceptrons-Graph Neural Network Algorithm for Protein Stability Prediction in Single- and Multiple-Point Mutations with Experimental Validation.Research (Wash D C). 2025 Apr 15;8:0674. doi: 10.34133/research.0674. eCollection 2025. Research (Wash D C). 2025. PMID: 40235597 Free PMC article.
-
Progress and challenges for the application of machine learning for neglected tropical diseases.F1000Res. 2025 Jul 17;12:287. doi: 10.12688/f1000research.129064.3. eCollection 2023. F1000Res. 2025. PMID: 40642109 Free PMC article. Review.
References
-
- Rebsamen M., Kandasamy R.K., Superti-Furga G.. Protein interaction networks in innate immunity. Trends Immunol. 2013; 34:610–619. - PubMed
-
- Lorch M., Mason J.M., Clarke A.R., Parker M.J.. Effects of core mutations on the folding of a β-sheet protein: implications for backbone organization in the I-state. Biochemistry. 1999; 38:1377–1385. - PubMed
-
- Lorch M., Mason J.M., Sessions R.B., Clarke A.R.. Effects of mutations on the thermodynamics of a protein folding reaction: implications for the mechanism of formation of the intermediate and transition states. Biochemistry. 2000; 39:3480–3485. - PubMed
-
- Alfalah M., Keiser M., Leeb T., Zimmer K.-P., Naim H.Y.. Compound heterozygous mutations affect protein folding and function in patients with congenital sucrase-isomaltase deficiency. Gastroenterology. 2009; 136:883–892. - PubMed

