Mild drought in the vegetative stage induces phenotypic, gene expression, and DNA methylation plasticity in Arabidopsis but no transgenerational effects
- PMID: 32166321
- PMCID: PMC7307858
- DOI: 10.1093/jxb/eraa132
Mild drought in the vegetative stage induces phenotypic, gene expression, and DNA methylation plasticity in Arabidopsis but no transgenerational effects
Abstract
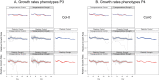
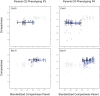
There is renewed interest in whether environmentally induced changes in phenotypes can be heritable. In plants, heritable trait variation can occur without DNA sequence mutations through epigenetic mechanisms involving DNA methylation. However, it remains unknown whether this alternative system of inheritance responds to environmental changes and if it can provide a rapid way for plants to generate adaptive heritable phenotypic variation. To assess potential transgenerational effects induced by the environment, we subjected four natural accessions of Arabidopsis thaliana together with the reference accession Col-0 to mild drought in a multi-generational experiment. As expected, plastic responses to drought were observed in each accession, as well as a number of intergenerational effects of the parental environments. However, after an intervening generation without stress, except for a very few trait-based parental effects, descendants of stressed and non-stressed plants were phenotypically indistinguishable irrespective of whether they were grown in control conditions or under water deficit. In addition, genome-wide analysis of DNA methylation and gene expression in Col-0 demonstrated that, while mild drought induced changes in the DNA methylome of exposed plants, these variants were not inherited. We conclude that mild drought stress does not induce transgenerational epigenetic effects.
Keywords: Arabidopsis; drought; epigenetics; maternal effects; methylation; plasticity; transgenerational effects.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures



Similar articles
-
Pre-conditioning the epigenetic response to high vapor pressure deficit increases the drought tolerance of Arabidopsis thaliana.Plant Signal Behav. 2013 Oct;8(10):e25974. doi: 10.4161/psb.25974. Plant Signal Behav. 2013. PMID: 24270688 Free PMC article.
-
Environmental heat and salt stress induce transgenerational phenotypic changes in Arabidopsis thaliana.PLoS One. 2013 Apr 9;8(4):e60364. doi: 10.1371/journal.pone.0060364. Print 2013. PLoS One. 2013. PMID: 23585834 Free PMC article.
-
DNA methylation mediates genetic variation for adaptive transgenerational plasticity.Proc Biol Sci. 2016 Sep 14;283(1838):20160988. doi: 10.1098/rspb.2016.0988. Proc Biol Sci. 2016. PMID: 27629032 Free PMC article.
-
Transgenerational inheritance: how impacts to the epigenetic and genetic information of parents affect offspring health.Hum Reprod Update. 2019 Sep 11;25(5):518-540. doi: 10.1093/humupd/dmz017. Hum Reprod Update. 2019. PMID: 31374565
-
Plant Transgenerational Epigenetics.Annu Rev Genet. 2016 Nov 23;50:467-491. doi: 10.1146/annurev-genet-120215-035254. Epub 2016 Oct 6. Annu Rev Genet. 2016. PMID: 27732791 Review.
Cited by
-
Establishment, maintenance, and biological roles of non-CG methylation in plants.Essays Biochem. 2019 Dec 20;63(6):743-755. doi: 10.1042/EBC20190032. Essays Biochem. 2019. PMID: 31652316 Free PMC article. Review.
-
A high-quality haplotype genome of Michelia alba DC reveals differences in methylation patterns and flower characteristics.Mol Hortic. 2024 May 29;4(1):23. doi: 10.1186/s43897-024-00098-z. Mol Hortic. 2024. PMID: 38807235 Free PMC article.
-
Genetic and Epigenetic Changes in Arabidopsis thaliana Exposed to Ultraviolet-C Radiation Stress for 25 Generations.Life (Basel). 2025 Mar 20;15(3):502. doi: 10.3390/life15030502. Life (Basel). 2025. PMID: 40141846 Free PMC article.
-
Photodamage repair pathways contribute to the accurate maintenance of the DNA methylome landscape upon UV exposure.PLoS Genet. 2019 Nov 18;15(11):e1008476. doi: 10.1371/journal.pgen.1008476. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31738755 Free PMC article.
-
Stress memory and its regulation in plants experiencing recurrent drought conditions.Theor Appl Genet. 2023 Feb 15;136(2):26. doi: 10.1007/s00122-023-04313-1. Theor Appl Genet. 2023. PMID: 36788199 Free PMC article. Review.
References
-
- Becker C, Hagmann J, Müller J, Koenig D, Stegle O, Borgwardt K, Weigel D. 2011. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249. - PubMed
-
- Biernacki C, Celeux G, Govaert G. 2000. Assessing a mixture model for clustering with the integrated completed likelihood. IEEE Transactions on Pattern Analysis and Machine Intelligence 22, 719–725.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

