Deep Neural Networks for Survival Analysis Using Pseudo Values
- PMID: 32167918
- PMCID: PMC8056290
- DOI: 10.1109/JBHI.2020.2980204
Deep Neural Networks for Survival Analysis Using Pseudo Values
Abstract
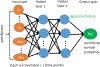
There has been increasing interest in modelling survival data using deep learning methods in medical research. Current approaches have focused on designing special cost functions to handle censored survival data. We propose a very different method with two simple steps. In the first step, we transform each subject's survival time into a series of jackknife pseudo conditional survival probabilities and then use these pseudo probabilities as a quantitative response variable in the deep neural network model. By using the pseudo values, we reduce a complex survival analysis to a standard regression problem, which greatly simplifies the neural network construction. Our two-step approach is simple, yet very flexible in making risk predictions for survival data, which is very appealing from the practice point of view. The source code is freely available at http://github.com/lilizhaoUM/DNNSurv.
Figures


References
-
- Martinsson E, “WTTE-RNN: Weibull time to event recurrent neural network,” Master’s thesis, University of Gothenburg, Sweden, 2016.
-
- Xinliang Zhu JY and Huang J, “Deep convolutional neural network for survival analysis with pathological images,” IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2016.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

