High-Speed Super-Resolution Imaging Using Protein-Assisted DNA-PAINT
- PMID: 32168456
- PMCID: PMC7146856
- DOI: 10.1021/acs.nanolett.9b04277
High-Speed Super-Resolution Imaging Using Protein-Assisted DNA-PAINT
Abstract
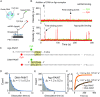
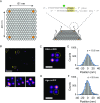
Super-resolution imaging allows for the visualization of cellular structures on a nanoscale level. DNA-PAINT (DNA point accumulation in nanoscale topology) is a super-resolution method that depends on the binding and unbinding of DNA imager strands. The current DNA-PAINT technique suffers from slow acquisition due to the low binding rate of the imager strands. Here we report on a method where imager strands are loaded into a protein, Argonaute (Ago), which allows for faster binding. Ago preorders the DNA imager strand into a helical conformation, allowing for 10 times faster target binding. Using a 2D DNA origami structure, we demonstrate that Ago-assisted DNA-PAINT (Ago-PAINT) can speed up the current DNA-PAINT technique by an order of magnitude, while maintaining the high spatial resolution. We envision this tool to be useful for super-resolution imaging and other techniques that rely on nucleic acid interactions.
Keywords: Ago-PAINT; Argonaute; DNA origami; DNA-PAINT; single-molecule FRET; super-resolution microscopy.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

