Genetically Modified Micro-Organisms for Industrial Food Enzyme Production: An Overview
- PMID: 32168815
- PMCID: PMC7143438
- DOI: 10.3390/foods9030326
Genetically Modified Micro-Organisms for Industrial Food Enzyme Production: An Overview
Abstract
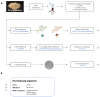
The use of food enzymes (FE) by the industrial food industry is continuously increasing. These FE are mainly obtained by microbial fermentation, for which both wild-type (WT) and genetically modified (GM) strains are used. The FE production yield can be increased by optimizing the fermentation process, either by using genetically modified micro-organism (GMM) strains or by producing recombinant enzymes. This review provides a general overview of the different methods used to produce FE preparations and how the use of GMM can increase the production yield. Additionally, information regarding the construction of these GMM strains is provided. Thereafter, an overview of the different European regulations concerning the authorization of FE preparations on the European market and the use of GMM strains is given. Potential issues related to the authorization and control of FE preparations sold on the European market are then identified and illustrated by a case study. This process highlighted the importance for control of FE preparations and the consequent need for appropriate detection methods targeting the presence of GMM, which is used in fermentation products.
Keywords: European regulations; food enzymes; genetically modified micro-organisms; safety control.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Industrial Enzymes Market by Type (Amylases, Cellulases, Proteases, Lipases, and Phytases), Application (Food & Beverages, Cleaning Agents, and Animal Feed), Source (Microorganism, Plant, and Animal), and Region—Global Forecast to 2022. Market Research Report. [(accessed on 6 January 2019)];2016 Available online: https://www.marketsandmarkets.com/Market-Reports/industrial-enzymes-mark....
-
- Chapman J., Ismail A.E., Dinu C.Z. Industrial applications of enzymes: Recent advances, techniques, and outlooks. Catalysts. 2018;8:238. doi: 10.3390/catal8060238. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

