Profiling the Salivary microbiome of the Qatari population
- PMID: 32169076
- PMCID: PMC7071716
- DOI: 10.1186/s12967-020-02291-2
Profiling the Salivary microbiome of the Qatari population
Abstract
Background: The role of the human microbiome in human health and disease has been studied in various body sites. However, compared to the gut microbiome, where most of the research focus is, the salivary microbiome still bears a vast amount of information that needs to be revealed. This study aims to characterize the salivary microbiome composition in the Qatari population, and to explore specific microbial signatures that can be associated with various lifestyles and different oral conditions.
Materials and methods: We characterized the salivary microbiome of 997 Qatari adults using high-throughput sequencing of the V1-V3 region of the 16S rRNA gene.
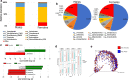
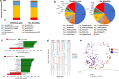
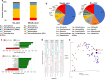
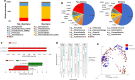
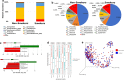
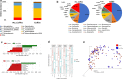
Results: In this study, we have characterized the salivary microbiome of 997 Qatari participants. Our data show that Bacteroidetes, Firmicutes, Actinobacteria and Proteobacteria are the common phyla isolated from the saliva samples, with Bacteroidetes being the most predominant phylum. Bacteroidetes was also more predominant in males versus females in the study cohort, although differences in the microbial diversity were not statistically significant. We also show that, a lower diversity of the salivary microbiome is observed in the elderly participants, with Prevotella and Treponema being the most significant genera. In participants with oral conditions such as mouth ulcers, bleeding or painful gum, our data show that Prevotella and Capnocytophaga are the most dominant genera as compared to the controls. Similar patterns were observed in participants with various smoking habits as compared to the non-smoking participants. Our data show that Streptococcus and Neisseria are more dominant among denture users, as compared to the non-denture users. Our data also show that, abnormal oral conditions are associated with a reduced microbial diversity and microbial richness. Moreover, in this study we show that frequent coffee drinkers have higher microbial diversity compared to the non-drinkers, indicating that coffee may cause changes to the salivary microbiome. Furthermore, tea drinkers show higher microbial richness as compared to the non-tea drinkers.
Conclusion: This is the first study to assess the salivary microbiome in an Arab population, and one of the largest population-based studies aiming to the characterize the salivary microbiome composition and its association with age, oral health, denture use, smoking and coffee-tea consumption.
Keywords: 16S rRNA gene sequencing; Dysbiosis; Oral health; Qatar Biobank; Qatari; Saliva.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


Similar articles
-
Smoking induced salivary microbiome dysbiosis and is correlated with lipid biomarkers.BMC Oral Health. 2024 May 25;24(1):608. doi: 10.1186/s12903-024-04340-4. BMC Oral Health. 2024. PMID: 38796419 Free PMC article.
-
Salivary Microbiome and Cigarette Smoking: A First of Its Kind Investigation in Jordan.Int J Environ Res Public Health. 2019 Dec 30;17(1):256. doi: 10.3390/ijerph17010256. Int J Environ Res Public Health. 2019. PMID: 31905907 Free PMC article.
-
High-throughput sequencing identifies salivary microbiota in Chinese caries-free preschool children with primary dentition.J Zhejiang Univ Sci B. 2021 Apr 15;22(4):285-294. doi: 10.1631/jzus.B2000554. J Zhejiang Univ Sci B. 2021. PMID: 33835762 Free PMC article.
-
The oral microbiome and human health.J Oral Sci. 2017;59(2):201-206. doi: 10.2334/josnusd.16-0856. J Oral Sci. 2017. PMID: 28637979 Review.
-
Insights into the human oral microbiome.Arch Microbiol. 2018 May;200(4):525-540. doi: 10.1007/s00203-018-1505-3. Epub 2018 Mar 23. Arch Microbiol. 2018. PMID: 29572583 Review.
Cited by
-
The association between denture use and cardiovascular diseases. The United States National Health and Nutrition Examination Survey 2009-2018.Front Cardiovasc Med. 2023 Jan 10;9:1000478. doi: 10.3389/fcvm.2022.1000478. eCollection 2022. Front Cardiovasc Med. 2023. PMID: 36704477 Free PMC article.
-
Salivary microbiome and hypertension in the Qatari population.J Transl Med. 2023 Jul 8;21(1):454. doi: 10.1186/s12967-023-04247-8. J Transl Med. 2023. PMID: 37422685 Free PMC article.
-
Comparative analysis of the human microbiome from four different regions of China and machine learning-based geographical inference.mSphere. 2025 Jan 28;10(1):e0067224. doi: 10.1128/msphere.00672-24. Epub 2024 Dec 19. mSphere. 2025. PMID: 39699186 Free PMC article.
-
Trick or Treating in Forensics-The Challenge of the Saliva Microbiome: A Narrative Review.Microorganisms. 2020 Sep 29;8(10):1501. doi: 10.3390/microorganisms8101501. Microorganisms. 2020. PMID: 33003446 Free PMC article. Review.
-
The oral microbiota and cardiometabolic health: A comprehensive review and emerging insights.Front Immunol. 2022 Nov 18;13:1010368. doi: 10.3389/fimmu.2022.1010368. eCollection 2022. Front Immunol. 2022. PMID: 36466857 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

