Aberrant Expression Profiles of lncRNAs and Their Associated Nearby Coding Genes in the Hippocampus of the SAMP8 Mouse Model with AD
- PMID: 32169802
- PMCID: PMC7066064
- DOI: 10.1016/j.omtn.2020.02.008
Aberrant Expression Profiles of lncRNAs and Their Associated Nearby Coding Genes in the Hippocampus of the SAMP8 Mouse Model with AD
Abstract
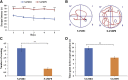
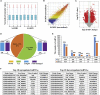
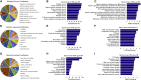
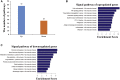
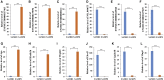
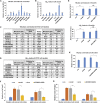
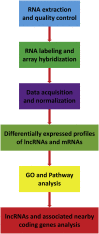
The senescence-accelerated mouse prone 8 (SAMP8) mouse model is a useful model for investigating the fundamental mechanisms involved in the age-related learning and memory deficits of Alzheimer's disease (AD), while the SAM/resistant 1 (SAMR1) mouse model shows normal features. Recent evidence has shown that long non-coding RNAs (lncRNAs) may play an important role in AD pathogenesis. However, a comprehensive and systematic understanding of the function of AD-related lncRNAs and their associated nearby coding genes in AD is still lacking. In this study, we collected the hippocampus, the main area of AD pathological processes, of SAMP8 and SAMR1 animals and performed microarray analysis to identify aberrantly expressed lncRNAs and their associated nearby coding genes, which may contribute to AD pathogenesis. We identified 3,112 differentially expressed lncRNAs and 3,191 differentially expressed mRNAs in SAMP8 mice compared to SAMR1 mice. More than 70% of the deregulated lncRNAs were intergenic and exon sense-overlapping lncRNAs. Gene Ontology (GO) and pathway analyses of the AD-related transcripts were also performed and are described in detail, which imply that metabolic process reprograming was likely related to AD. Furthermore, six lncRNAs and six mRNAs were selected for further validation of the microarray results using quantitative PCR, and the results were consistent with the findings from the microarray. Moreover, we analyzed 780 lincRNAs (also called long "intergenic" non-coding RNAs) and their associated nearby coding genes. Among these lincRNAs, AK158400 had the most genes nearby (n = 13), all of which belonged to the histone cluster 1 family, suggesting regulation of the nucleosome structure of the chromosomal fiber by affecting nearby genes during AD progression. In addition, we also identified 97 aberrant antisense lncRNAs and their associated coding genes. It is likely that these dysregulated lncRNAs and their associated nearby coding genes play a role in the development and/or progression of AD.
Keywords: Alzheimer’s disease; lncRNA-associated nearby genes; lncRNAs.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Yaari R., Corey-Bloom J. Alzheimer’s disease. Semin. Neurol. 2007;27:32–41. - PubMed
-
- Jaroudi W., Garami J., Garrido S., Hornberger M., Keri S., Moustafa A.A. Factors underlying cognitive decline in old age and Alzheimer’s disease: the role of the hippocampus. Rev. Neurosci. 2017;28:705–714. - PubMed
-
- Anand R., Gill K.D., Mahdi A.A. Therapeutics of Alzheimer’s disease: past, present and future. Neuropharmacology. 2014;76:27–50. - PubMed
-
- Zhang C., Du Q.Y., Chen L.D., Wu W.H., Liao S.Y., Yu L.H., Liang X.T. Design, synthesis and evaluation of novel tacrine-multialkoxybenzene hybrids as multi-targeted compounds against Alzheimer’s disease. Eur. J. Med. Chem. 2016;116:200–209. - PubMed
LinkOut - more resources
Full Text Sources

