The transcriptional regulator MEIS2 sets up the ground state for palatal osteogenesis in mice
- PMID: 32169905
- PMCID: PMC7170518
- DOI: 10.1074/jbc.RA120.012684
The transcriptional regulator MEIS2 sets up the ground state for palatal osteogenesis in mice
Abstract
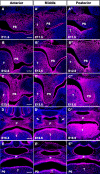
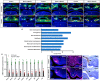
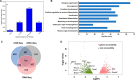
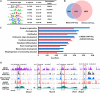
Haploinsufficiency of Meis homeobox 2 (MEIS2), encoding a transcriptional regulator, is associated with human cleft palate, and Meis2 inactivation leads to abnormal palate development in mice, implicating MEIS2 functions in palate development. However, its functional mechanisms remain unknown. Here we observed widespread MEIS2 expression in the developing palate in mice. Wnt1Cre -mediated Meis2 inactivation in cranial neural crest cells led to a secondary palate cleft. Importantly, about half of the Wnt1Cre ;Meis2f/f mice exhibited a submucous cleft, providing a model for studying palatal bone formation and patterning. Consistent with complete absence of palatal bones, the results from integrative analyses of MEIS2 by ChIP sequencing, RNA-Seq, and an assay for transposase-accessible chromatin sequencing identified key osteogenic genes regulated directly by MEIS2, indicating that it plays a fundamental role in palatal osteogenesis. De novo motif analysis uncovered that the MEIS2-bound regions are highly enriched in binding motifs for several key osteogenic transcription factors, particularly short stature homeobox 2 (SHOX2). Comparative ChIP sequencing analyses revealed genome-wide co-occupancy of MEIS2 and SHOX2 in addition to their colocalization in the developing palate and physical interaction, suggesting that SHOX2 and MEIS2 functionally interact. However, although SHOX2 was required for proper palatal bone formation and was a direct downstream target of MEIS2, Shox2 overexpression failed to rescue the palatal bone defects in a Meis2-mutant background. These results, together with the fact that Meis2 expression is associated with high osteogenic potential and required for chromatin accessibility of osteogenic genes, support a vital function of MEIS2 in setting up a ground state for palatal osteogenesis.
Keywords: Meis homeobox 2 (MEIS2); bone; chromatin accessibility; craniofacial development; gene knockout; gene regulation; osteogenesis; palatal development; short stature homeobox 2 (SHOX2); transcription; transcription factor; transgenic mice.
© 2020 Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures


Similar articles
-
Shox2 regulates osteogenic differentiation and pattern formation during hard palate development in mice.J Biol Chem. 2019 Nov 29;294(48):18294-18305. doi: 10.1074/jbc.RA119.008801. Epub 2019 Oct 24. J Biol Chem. 2019. PMID: 31649032 Free PMC article.
-
Neural crest-specific deletion of Ldb1 leads to cleft secondary palate with impaired palatal shelf elevation.BMC Dev Biol. 2014 Jan 17;14:3. doi: 10.1186/1471-213X-14-3. BMC Dev Biol. 2014. PMID: 24433583 Free PMC article.
-
A unique stylopod patterning mechanism by Shox2-controlled osteogenesis.Development. 2016 Jul 15;143(14):2548-60. doi: 10.1242/dev.138750. Epub 2016 Jun 10. Development. 2016. PMID: 27287812 Free PMC article.
-
Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate.Development. 2005 Oct;132(19):4397-406. doi: 10.1242/dev.02013. Epub 2005 Sep 1. Development. 2005. PMID: 16141225
-
Regional regulation of palatal growth and patterning along the anterior-posterior axis in mice.J Anat. 2005 Nov;207(5):655-67. doi: 10.1111/j.1469-7580.2005.00474.x. J Anat. 2005. PMID: 16313398 Free PMC article. Review.
Cited by
-
Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum.Development. 2022 Feb 15;149(4):dev200035. doi: 10.1242/dev.200035. Epub 2022 Feb 23. Development. 2022. PMID: 35156680 Free PMC article.
-
Single cell sequencing of the mouse anterior palate reveals mesenchymal heterogeneity.Dev Dyn. 2023 Jun;252(6):713-727. doi: 10.1002/dvdy.573. Epub 2023 Feb 23. Dev Dyn. 2023. PMID: 36734036 Free PMC article.
-
Osteogenic and angiogenic profiles of the palatal process of the maxilla and the palatal process of the palatine bone.J Anat. 2022 Feb;240(2):385-397. doi: 10.1111/joa.13545. Epub 2021 Sep 26. J Anat. 2022. PMID: 34569061 Free PMC article.
-
Cleft palate, congenital heart disease, and developmental delay involving MEIS2 heterozygous mutations found in the patient with attention deficit hyperactivity disorder: a case report.Front Pediatr. 2024 Dec 24;12:1500152. doi: 10.3389/fped.2024.1500152. eCollection 2024. Front Pediatr. 2024. PMID: 39776641 Free PMC article.
-
MEIS2 (15q14) gene deletions in siblings with mild developmental phenotypes and bifid uvula: documentation of mosaicism in an unaffected parent.Mol Cytogenet. 2021 Dec 20;14(1):58. doi: 10.1186/s13039-021-00570-1. Mol Cytogenet. 2021. PMID: 34930369 Free PMC article.
References
-
- Dibner C., Elias S., and Frank D. (2001) XMeis3 protein activity is required for proper hindbrain patterning in Xenopus laevis embryos. Development 128, 3415–3426 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

