A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice
- PMID: 32169907
- PMCID: PMC7569398
- DOI: 10.1136/gutjnl-2019-320105
A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice
Abstract
Objective: Gut microbiota have been linked to inflammatory bowel disease (IBD) and colorectal cancer (CRC). Akkermansia muciniphila (A. muciniphila) is a gram-negative anaerobic bacterium that is selectively decreased in the faecal microbiota of patients with IBD, but its causative role and molecular mechanism in blunting colitis-associated colorectal cancer (CAC) remain inconclusive. This study investigates how A. muciniphila engages the immune response in CAC.
Design: Mice were given dextran sulfate sodium to induce colitis, followed by azoxymethane to establish CAC with or without pasteurised A. muciniphila or a specific outer membrane protein (Amuc_1100) treatment. Faeces from mice and patients with IBD or CRC were collected for 16S rRNA sequencing. The effects of A. muciniphila or Amuc_1100 on the immune response in acute colitis and CAC were investigated.
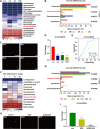
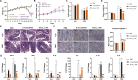
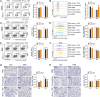
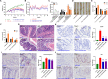
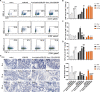
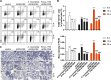
Results: A. muciniphila was significantly reduced in patients with IBD and mice with colitis or CAC. A. muciniphila or Amuc_1100 could improve colitis, with a reduction in infiltrating macrophages and CD8+ cytotoxic T lymphocytes (CTLs) in the colon. Their treatment also decreased CD16/32+ macrophages in the spleen and mesenteric lymph nodes (MLN) of colitis mice. Amuc_1100 elevated PD-1+ CTLs in the spleen. Moreover, A. muciniphila and Amuc_1100 blunted tumourigenesis by expanding CTLs in the colon and MLN. Remarkably, they activated CTLs in the MLN, as indicated by TNF-α induction and PD-1downregulation. Amuc_1100 could stimulate and activate CTLs from splenocytes in CT26 cell conditioned medium.
Conclusions: These data indicate that pasteurised A. muciniphila or Amuc_1100 can blunt colitis and CAC through the modulation of CTLs.
Keywords: colonic microflora; colorectal cancer; experimental colitis; gut immunology; nutritional supplementation.
© Author(s) (or their employer(s)) 2020. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
