NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture
- PMID: 32170015
- PMCID: PMC7132277
- DOI: 10.1073/pnas.1909546117
NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture
Abstract
Neutrophil extracellular traps (NETs) are web-like DNA structures decorated with histones and cytotoxic proteins that are released by activated neutrophils to trap and neutralize pathogens during the innate immune response, but also form in and exacerbate sterile inflammation. Peptidylarginine deiminase 4 (PAD4) citrullinates histones and is required for NET formation (NETosis) in mouse neutrophils. While the in vivo impact of NETs is accumulating, the cellular events driving NETosis and the role of PAD4 in these events are unclear. We performed high-resolution time-lapse microscopy of mouse and human neutrophils and differentiated HL-60 neutrophil-like cells (dHL-60) labeled with fluorescent markers of organelles and stimulated with bacterial toxins or Candida albicans to induce NETosis. Upon stimulation, cells exhibited rapid disassembly of the actin cytoskeleton, followed by shedding of plasma membrane microvesicles, disassembly and remodeling of the microtubule and vimentin cytoskeletons, ER vesiculation, chromatin decondensation and nuclear rounding, progressive plasma membrane and nuclear envelope (NE) permeabilization, nuclear lamin meshwork and then NE rupture to release DNA into the cytoplasm, and finally plasma membrane rupture and discharge of extracellular DNA. Inhibition of actin disassembly blocked NET release. Mouse and dHL-60 cells bearing genetic alteration of PAD4 showed that chromatin decondensation, lamin meshwork and NE rupture and extracellular DNA release required the enzymatic and nuclear localization activities of PAD4. Thus, NETosis proceeds by a stepwise sequence of cellular events culminating in the PAD4-mediated expulsion of DNA.
Keywords: innate immunity; microscopy; neutrophil.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


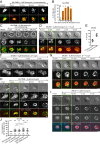
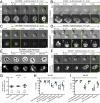


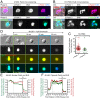

Comment in
-
Functional actin cytoskeleton is required in early stage of NETosis induction.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):22653-22654. doi: 10.1073/pnas.2013790117. Proc Natl Acad Sci U S A. 2020. PMID: 32943582 Free PMC article. No abstract available.
-
Reply to Liu: The disassembly of the actin cytoskeleton is an early event during NETosis.Proc Natl Acad Sci U S A. 2020 Sep 15;117(37):22655-22656. doi: 10.1073/pnas.2015951117. Proc Natl Acad Sci U S A. 2020. PMID: 32943583 Free PMC article. No abstract available.
Similar articles
-
Citrullination Licenses Calpain to Decondense Nuclei in Neutrophil Extracellular Trap Formation.Front Immunol. 2019 Oct 22;10:2481. doi: 10.3389/fimmu.2019.02481. eCollection 2019. Front Immunol. 2019. PMID: 31695698 Free PMC article.
-
Protein Deiminase 4 and CR3 Regulate Aspergillus fumigatus and β-Glucan-Induced Neutrophil Extracellular Trap Formation, but Hyphal Killing Is Dependent Only on CR3.Front Immunol. 2018 May 29;9:1182. doi: 10.3389/fimmu.2018.01182. eCollection 2018. Front Immunol. 2018. PMID: 29896200 Free PMC article.
-
Neutrophil elastase-deficient mice form neutrophil extracellular traps in an experimental model of deep vein thrombosis.J Thromb Haemost. 2016 Mar;14(3):551-8. doi: 10.1111/jth.13239. Epub 2016 Feb 5. J Thromb Haemost. 2016. PMID: 26712312 Free PMC article.
-
Cellular Mechanisms of NETosis.Annu Rev Cell Dev Biol. 2020 Oct 6;36:191-218. doi: 10.1146/annurev-cellbio-020520-111016. Epub 2020 Jul 14. Annu Rev Cell Dev Biol. 2020. PMID: 32663035 Free PMC article. Review.
-
[Progress in mechanism of formation of neutrophil extracellular traps: Review].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020 Jun;36(6):561-564. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020. PMID: 32696748 Review. Chinese.
Cited by
-
Dysregulation of neutrophil death in sepsis.Front Immunol. 2022 Aug 18;13:963955. doi: 10.3389/fimmu.2022.963955. eCollection 2022. Front Immunol. 2022. PMID: 36059483 Free PMC article. Review.
-
PAD4 takes charge during neutrophil activation: Impact of PAD4 mediated NET formation on immune-mediated disease.J Thromb Haemost. 2021 Jul;19(7):1607-1617. doi: 10.1111/jth.15313. Epub 2021 May 12. J Thromb Haemost. 2021. PMID: 33773016 Free PMC article. Review.
-
Cyclic stretch enhances neutrophil extracellular trap formation.BMC Biol. 2024 Sep 18;22(1):209. doi: 10.1186/s12915-024-02009-6. BMC Biol. 2024. PMID: 39289752 Free PMC article.
-
Tyrosine phosphatase SHP2 exacerbates psoriasis-like skin inflammation in mice via ERK5-dependent NETosis.MedComm (2020). 2022 Mar 4;3(1):e120. doi: 10.1002/mco2.120. eCollection 2022 Mar. MedComm (2020). 2022. PMID: 35281792 Free PMC article.
-
CD44/ERM/F-actin complex mediates targeted nuclear degranulation and excessive neutrophil extracellular trap formation during sepsis.J Cell Mol Med. 2022 Apr;26(7):2089-2103. doi: 10.1111/jcmm.17231. Epub 2022 Feb 11. J Cell Mol Med. 2022. PMID: 35146909 Free PMC article.
References
-
- Brinkmann V., et al. , Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004). - PubMed
-
- Urban C. F., Reichard U., Brinkmann V., Zychlinsky A., Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 8, 668–676 (2006). - PubMed
-
- Saitoh T., et al. , Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12, 109–116 (2012). - PubMed
-
- Clark S. R., et al. , Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13, 463–469 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

