Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization
- PMID: 32170131
- PMCID: PMC7070081
- DOI: 10.1038/s41467-020-15165-4
Dynamics of bacterial population growth in biofilms resemble spatial and structural aspects of urbanization
Abstract
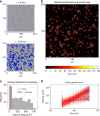
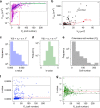
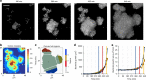
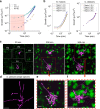
Biofilms develop from bacteria bound on surfaces that grow into structured communities (microcolonies). Although surface topography is known to affect bacterial colonization, how multiple individual settlers develop into microcolonies simultaneously remains underexplored. Here, we use multiscale population-growth and 3D-morphometric analyses to assess the spatiotemporal development of hundreds of bacterial colonizers towards submillimeter-scale microcolony communities. Using an oral bacterium (Streptococcus mutans), we find that microbial cells settle on the surface randomly under sucrose-rich conditions, regardless of surface topography. However, only a subset of colonizers display clustering behavior and growth following a power law. These active colonizers expand three-dimensionally by amalgamating neighboring bacteria into densely populated microcolonies. Clustering and microcolony assembly are dependent on exopolysaccharides, while population growth dynamics and spatial structure are affected by cooperative or antagonistic microbes. Our work suggests that biofilm assembly resembles certain spatial-structural features of urbanization, where population growth and expansion can be influenced by type of settlers, neighboring cells, and further community merging and scaffolding occurring at various scales.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Wimpenny, J., Manz, W. & Szewzyk, U. Heterogeneity in biofilms. FEMS Microbiol. Rev.10.1016/S0168-6445(00)00052-8 (2000). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

