Tead transcription factors differentially regulate cortical development
- PMID: 32170161
- PMCID: PMC7070074
- DOI: 10.1038/s41598-020-61490-5
Tead transcription factors differentially regulate cortical development
Abstract
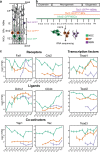
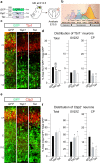
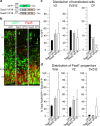
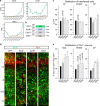
Neural stem cells (NSCs) generate neurons of the cerebral cortex with distinct morphologies and functions. How specific neuron production, differentiation and migration are orchestrated is unclear. Hippo signaling regulates gene expression through Tead transcription factors (TFs). We show that Hippo transcriptional coactivators Yap1/Taz and the Teads have distinct functions during cortical development. Yap1/Taz promote NSC maintenance and Satb2+ neuron production at the expense of Tbr1+ neuron generation. However, Teads have moderate effects on NSC maintenance and do not affect Satb2+ neuron differentiation. Conversely, whereas Tead2 blocks Tbr1+ neuron formation, Tead1 and Tead3 promote this early fate. In addition, we found that Hippo effectors regulate neuronal migration to the cortical plate (CP) in a reciprocal fashion, that ApoE, Dab2 and Cyr61 are Tead targets, and these contribute to neuronal fate determination and migration. Our results indicate that multifaceted Hippo signaling is pivotal in different aspects of cortical development.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

