The Aurora B specificity switch is required to protect from non-disjunction at the metaphase/anaphase transition
- PMID: 32170202
- PMCID: PMC7070073
- DOI: 10.1038/s41467-020-15163-6
The Aurora B specificity switch is required to protect from non-disjunction at the metaphase/anaphase transition
Erratum in
-
Author Correction: The Aurora B specificity switch is required to protect from non-disjunction at the metaphase/anaphase transition.Nat Commun. 2020 Jun 3;11(1):2884. doi: 10.1038/s41467-020-16468-2. Nat Commun. 2020. PMID: 32493898 Free PMC article.
Abstract
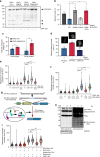
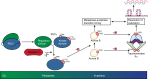
The Aurora B abscission checkpoint delays cytokinesis until resolution of DNA trapped in the cleavage furrow. This process involves PKCε phosphorylation of Aurora B S227. Assessing if this PKCε-Aurora B module provides a more widely exploited genome-protective control for the cell cycle, we show Aurora B phosphorylation at S227 by PKCε also occurs during mitosis. Expression of Aurora B S227A phenocopies inhibition of PKCε in by-passing the delay and resolution at anaphase entry that is associated with non-disjunction and catenation of sister chromatids. Implementation of this anaphase delay is reflected in PKCε activation following cell cycle dependent cleavage by caspase 7; knock-down of caspase 7 phenocopies PKCε loss, in a manner rescued by ectopically expressing/generating a free PKCε catalytic domain. Molecular dynamics indicates that Aurora B S227 phosphorylation induces conformational changes and this manifests in a profound switch in specificity towards S29 TopoIIα phosphorylation, a response necessary for catenation resolution during mitosis.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

