Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress
- PMID: 32171257
- PMCID: PMC7071642
- DOI: 10.1186/s12864-020-6622-0
Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress
Abstract
Background: Plants have developed various sophisticated mechanisms to cope up with climate extremes and different stress conditions, especially by involving specific transcription factors (TFs). The members of the WRKY TF family are well known for their role in plant development, phytohormone signaling and developing resistance against biotic or abiotic stresses. In this study, we performed a genome-wide screening to identify and analyze the WRKY TFs in pearl millet (Pennisetum glaucum; PgWRKY), which is one of the most widely grown cereal crops in the semi-arid regions.
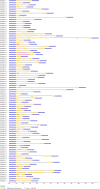
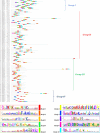
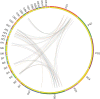
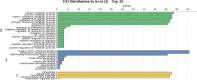
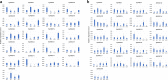
Results: A total number of 97 putative PgWRKY proteins were identified and classified into three major Groups (I-III) based on the presence of WRKY DNA binding domain and zinc-finger motif structures. Members of Group II have been further subdivided into five subgroups (IIa-IIe) based on the phylogenetic analysis. In-silico analysis of PgWRKYs revealed the presence of various cis-regulatory elements in their promoter region like ABRE, DRE, ERE, EIRE, Dof, AUXRR, G-box, etc., suggesting their probable involvement in growth, development and stress responses of pearl millet. Chromosomal mapping evidenced uneven distribution of identified 97 PgWRKY genes across all the seven chromosomes of pearl millet. Synteny analysis of PgWRKYs established their orthologous and paralogous relationship among the WRKY gene family of Arabidopsis thaliana, Oryza sativa and Setaria italica. Gene ontology (GO) annotation functionally categorized these PgWRKYs under cellular components, molecular functions and biological processes. Further, the differential expression pattern of PgWRKYs was noticed in different tissues (leaf, stem, root) and under both drought and salt stress conditions. The expression pattern of PgWRKY33, PgWRKY62 and PgWRKY65 indicates their probable involvement in both dehydration and salinity stress responses in pearl millet.
Conclusion: Functional characterization of identified PgWRKYs can be useful in delineating their role behind the natural stress tolerance of pearl millet against harsh environmental conditions. Further, these PgWRKYs can be employed in genome editing for millet crop improvement.
Keywords: Abiotic stress; Cis-regulatory elements; Pearl millet; Synteny; WRKY transcription factors.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Parry M, Arnell N, McMichael T, Nicholls R, Martens P, Kovats S, Livermore M, Rosenzweig C, Iglesias A, Fischer G. Millions at risk: defining critical climate change threats and targets. Glob Environ Chang. 2001;11(3):181–183.
-
- Ziervogel G, New M, Archer van Garderen E, Midgley G, Taylor A, Hamann R, Stuart-Hill S, Myers J, Warburton M. Climate change impacts and adaptation in South Africa. Wiley Interdiscip Rev Clim Chang. 2014;5(5):605–620.
-
- Beddington JR, Asaduzzaman M, Fernandez A, Clark ME, Guillou M, Jahn MM, Erda L, Mamo T, Bo N, Nobre CA. Achieving food security in the face of climate change: final report from the commission on sustainable agriculture and climate change. 2012.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

