One-step RT-qPCR assay for ZIKV RNA detection in Aedes aegypti samples: a protocol to study infection and gene expression during ZIKV infection
- PMID: 32171303
- PMCID: PMC7071672
- DOI: 10.1186/s13071-020-4002-x
One-step RT-qPCR assay for ZIKV RNA detection in Aedes aegypti samples: a protocol to study infection and gene expression during ZIKV infection
Abstract
Background: Zika virus (ZIKV) is transmitted to humans during the bite of an infected mosquito. In a scenario of globalization and climate change, the frequency of outbreaks has and will increase in areas with competent vectors, revealing a need for continuous improvement of ZIKV detection tools in vector populations. A simple, rapid and sensitive assay for viral detection is quantitative reverse transcription polymerase chain reaction (qRT-PCR), yet oligos optimized for ZIKV detection in mammalian cells and samples have repeatedly shown high background when used on mosquito ribonucleic acid (RNA). In this paper, we present a one-step qRT-PCR protocol that allows for the detection of ZIKV in mosquitoes and for the evaluation of gene expression from the same mosquito sample and RNA. This assay is a less expensive qRT-PCR approach than that most frequently used in the literature and has a much lower background, allowing confident detection.
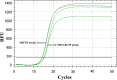
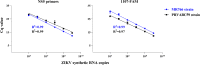
Methods: Our new oligo design to detect ZIKV RNA included in silico analysis of both viral and mosquito (Ae. aegypti and Ae. albopictus) genomes, targeting sequences conserved between Asian and African ZIKV lineages, but not matching Aedes genomes. This assay will allow researchers to avoid nonspecific amplification in insect samples due to viral integration into the mosquito genome, a phenomenon known to happen in wild and colonized populations of mosquitoes. Standard curves constructed with in vitro transcribed ZIKV RNA were used to optimize the sensitivity, efficiency and reproducibility of the assay.
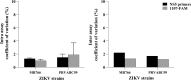
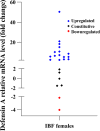
Results: Finally, the assay was used with success to detect both ZIKV RNA in infected mosquitoes and to detect expression of the Defensin A gene, an antimicrobial peptide (AMP) involved in Aedes aegypti immune response to virus infection.
Conclusions: The experimental approach to detect ZIKV RNA in Aedes aegypti presented here has demonstrated to be specific, sensitive and reliable, and additionally it allows for the analysis of mosquito gene expression during ZIKV infection.
Keywords: Aedes aegypti; Mosquito gene expression; One-step qRT-PCR; Zika virus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Development and Validation of Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) for Rapid Detection of ZIKV in Mosquito Samples from Brazil.Sci Rep. 2019 Mar 14;9(1):4494. doi: 10.1038/s41598-019-40960-5. Sci Rep. 2019. PMID: 30872672 Free PMC article.
-
SYBR green-based one step quantitative real-time polymerase chain reaction assay for the detection of Zika virus in field-caught mosquitoes.Parasit Vectors. 2017 Sep 19;10(1):427. doi: 10.1186/s13071-017-2373-4. Parasit Vectors. 2017. PMID: 28927458 Free PMC article.
-
Evidence for infection but not transmission of Zika virus by Aedes albopictus (Diptera: Culicidae) from Spain.Parasit Vectors. 2019 May 3;12(1):204. doi: 10.1186/s13071-019-3467-y. Parasit Vectors. 2019. PMID: 31053164 Free PMC article.
-
Aedes albopictus is a competent vector of Zika virus: A meta-analysis.PLoS One. 2019 May 21;14(5):e0216794. doi: 10.1371/journal.pone.0216794. eCollection 2019. PLoS One. 2019. PMID: 31112569 Free PMC article.
-
Zika Virus.Clin Microbiol Rev. 2016 Jul;29(3):487-524. doi: 10.1128/CMR.00072-15. Clin Microbiol Rev. 2016. PMID: 27029595 Free PMC article. Review.
Cited by
-
A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons.Genome Res. 2021 Mar;31(3):512-528. doi: 10.1101/gr.265157.120. Epub 2021 Jan 8. Genome Res. 2021. PMID: 33419731 Free PMC article.
-
A Single-Dose mRNA Vaccine Employing Porous Silica Nanoparticles Induces Robust Immune Responses Against the Zika Virus.Adv Sci (Weinh). 2024 Sep;11(35):e2404590. doi: 10.1002/advs.202404590. Epub 2024 Jul 15. Adv Sci (Weinh). 2024. PMID: 39010673 Free PMC article.
-
Dengue Virus-2 Infection Affects Fecundity and Elicits Specific Transcriptional Changes in the Ovaries of Aedes aegypti Mosquitoes.Front Microbiol. 2022 Jun 23;13:886787. doi: 10.3389/fmicb.2022.886787. eCollection 2022. Front Microbiol. 2022. PMID: 35814655 Free PMC article.
-
Temperate Conditions Limit Zika Virus Genome Replication.J Virol. 2022 May 25;96(10):e0016522. doi: 10.1128/jvi.00165-22. Epub 2022 Apr 25. J Virol. 2022. PMID: 35467365 Free PMC article.
References
-
- WHO Zika virus outbreaks in the Americas. Wkly Epidemiol Rec. 2015;90:609–610. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

