Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma
- PMID: 32173382
- PMCID: PMC8418904
- DOI: 10.1016/j.jhep.2020.03.008
Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma
Abstract
Background & aims: Cholangiocarcinoma (CCA), a deadly malignancy of the bile ducts, can be classified based on its anatomical location into either intrahepatic (iCCA) or extrahepatic (eCCA), each with different pathogenesis and clinical management. There is limited understanding of the molecular landscape of eCCA and no targeted therapy with clinical efficacy has been approved. We aimed to provide a molecular classification of eCCA and identify potential targets for molecular therapies.
Methods: An integrative genomic analysis of an international multicenter cohort of 189 eCCA cases was conducted. Genomic analysis included whole-genome expression, targeted DNA-sequencing and immunohistochemistry. Molecular findings were validated in an external set of 181 biliary tract tumors from the ICGC.
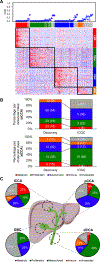
Results: KRAS (36.7%), TP53 (34.7%), ARID1A (14%) and SMAD4 (10.7%) were the most prevalent mutations, with ∼25% of tumors having a putative actionable genomic alteration according to OncoKB. Transcriptome-based unsupervised clustering helped us define 4 molecular classes of eCCA. Tumors classified within the Metabolic class (19%) showed a hepatocyte-like phenotype with activation of the transcription factor HNF4A and enrichment in gene signatures related to bile acid metabolism. The Proliferation class (23%), more common in patients with distal CCA, was characterized by enrichment of MYC targets, ERBB2 mutations/amplifications and activation of mTOR signaling. The Mesenchymal class (47%) was defined by signatures of epithelial-mesenchymal transition, aberrant TGFβ signaling and poor overall survival. Finally, tumors in the Immune class (11%) had a higher lymphocyte infiltration, overexpression of PD-1/PD-L1 and molecular features associated with a better response to immune checkpoint inhibitors.
Conclusion: An integrative molecular characterization identified distinct subclasses of eCCA. Genomic traits of each class provide the rationale for exploring patient stratification and novel therapeutic approaches.
Lay summary: Targeted therapies have not been approved for the treatment of extrahepatic cholangiocarcinoma. We performed a multi-platform molecular characterization of this tumor in a cohort of 189 patients. These analyses revealed 4 novel transcriptome-based molecular classes of extrahepatic cholangiocarcinoma and identified ∼25% of tumors with actionable genomic alterations, which has potential prognostic and therapeutic implications.
Keywords: Biomarkers; Extrahepatic cholangiocarcinoma; Immunotherapy; Liver cancer; Molecular classification; Targeted therapies.
Copyright © 2020 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest J.M.L. is receiving research support from Bayer HealthCare Pharmaceuticals, Eisai Inc, Bristol-Myers Squibb, Boehringer-Ingelheim and Ipsen, and consulting fees from Bayer HealthCare Pharmaceuticals, Merck, Eisai Inc, Bristol-Myers Squibb, Celsion Corporation, Eli Lilly, Roche, Genentech, Glycotest, Nucleix, Can-Fite Biopharma, AstraZeneca, and Exelixis. A.V. reports personal fees from NGM Pharmaceuticals, Gilead, Nucleix, Fuji Wako, Guidepoint and Exact Sciences. L.R.R. is receiving research support from Ariad Pharmaceuticals, Bayer, BTG International, Exact Sciences, Gilead Sciences, GRAIL Inc., RedHill Biopharma Ltd., TARGET PharmaSolutions and Wako Diagnostics, and is advisory board member of Bayer, Exact Sciences, Gilead Sciences, GRAIL, Inc., QED Therapeutics, Inc. and TAVEC. B.M. reports personal fees from Bayer and Gilead. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures




References
-
- Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, et al. Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat Rev Gastroenterol Hepatol 2016;13:261–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

