Empirically-derived synthetic populations to mitigate small sample sizes
- PMID: 32173502
- PMCID: PMC7839232
- DOI: 10.1016/j.jbi.2020.103408
Empirically-derived synthetic populations to mitigate small sample sizes
Abstract
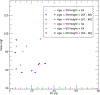
Limited sample sizes can lead to spurious modeling findings in biomedical research. The objective of this work is to present a new method to generate synthetic populations (SPs) from limited samples using matched case-control data (n = 180 pairs), considered as two separate limited samples. SPs were generated with multivariate kernel density estimations (KDEs) with unconstrained bandwidth matrices. We included four continuous variables and one categorical variable for each individual. Bandwidth matrices were determined with Differential Evolution (DE) optimization by covariance comparisons. Four synthetic samples (n = 180) were derived from their respective SPs. Similarity between observed samples with synthetic samples was compared assuming their empirical probability density functions (EPDFs) were similar. EPDFs were compared with the maximum mean discrepancy (MMD) test statistic based on the Kernel Two-Sample Test. To evaluate similarity within a modeling context, EPDFs derived from the Principal Component Analysis (PCA) scores and residuals were summarized with the distance to the model in X-space (DModX) as additional comparisons. Four SPs were generated from each sample. The probability of selecting a replicate when randomly constructing synthetic samples (n = 180) was infinitesimally small. MMD tests indicated that the observed sample EPDFs were similar to the respective synthetic EPDFs. For the samples, PCA scores and residuals did not deviate significantly when compared with their respective synthetic samples. The feasibility of this approach was demonstrated by producing synthetic data at the individual level, statistically similar to the observed samples. The methodology coupled KDE with DE optimization and deployed novel similarity metrics derived from PCA. This approach could be used to generate larger-sized synthetic samples. To develop this approach into a research tool for data exploration purposes, additional evaluation with increased dimensionality is required. Moreover, given a fully specified population, the degree to which individuals can be discarded while synthesizing the respective population accurately will be investigated. When these objectives are addressed, comparisons with other techniques such as bootstrapping will be required for a complete evaluation.
Keywords: Differential evolution; Distance to the model in X-space; Kernel density estimation; Overfitting; Principal component analysis; Synthetic data generation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


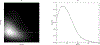

Similar articles
-
Techniques to produce and evaluate realistic multivariate synthetic data.Sci Rep. 2023 Jul 28;13(1):12266. doi: 10.1038/s41598-023-38832-0. Sci Rep. 2023. PMID: 37507387 Free PMC article.
-
Effect of finite sample size on feature selection and classification: a simulation study.Med Phys. 2010 Feb;37(2):907-20. doi: 10.1118/1.3284974. Med Phys. 2010. PMID: 20229900 Free PMC article.
-
Two-sample statistics based on anisotropic kernels.Inf inference. 2020 Sep;9(3):677-719. doi: 10.1093/imaiai/iaz018. Epub 2019 Dec 10. Inf inference. 2020. PMID: 32929389 Free PMC article.
-
Subgroup analyses in randomised controlled trials: quantifying the risks of false-positives and false-negatives.Health Technol Assess. 2001;5(33):1-56. doi: 10.3310/hta5330. Health Technol Assess. 2001. PMID: 11701102 Review.
-
Sample size calculation in metabolic phenotyping studies.Brief Bioinform. 2015 Sep;16(5):813-9. doi: 10.1093/bib/bbu052. Epub 2015 Jan 19. Brief Bioinform. 2015. PMID: 25600654 Review.
Cited by
-
A Simple-to-Use R Package for Mimicking Study Data by Simulations.Methods Inf Med. 2023 Sep;62(3-04):119-129. doi: 10.1055/a-2048-7692. Epub 2023 Mar 7. Methods Inf Med. 2023. PMID: 36882158 Free PMC article.
-
Synthetic Tabular Data Evaluation in the Health Domain Covering Resemblance, Utility, and Privacy Dimensions.Methods Inf Med. 2023 Jun;62(S 01):e19-e38. doi: 10.1055/s-0042-1760247. Epub 2023 Jan 9. Methods Inf Med. 2023. PMID: 36623830 Free PMC article.
-
Comparison of Machine Learning Techniques for Mortality Prediction in a Prospective Cohort of Older Adults.Int J Environ Res Public Health. 2021 Dec 4;18(23):12806. doi: 10.3390/ijerph182312806. Int J Environ Res Public Health. 2021. PMID: 34886532 Free PMC article.
-
Techniques to produce and evaluate realistic multivariate synthetic data.Sci Rep. 2023 Jul 28;13(1):12266. doi: 10.1038/s41598-023-38832-0. Sci Rep. 2023. PMID: 37507387 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

