Amygdala-Based Altered miRNome and Epigenetic Contribution of miR-128-3p in Conferring Susceptibility to Depression-Like Behavior via Wnt Signaling
- PMID: 32173733
- PMCID: PMC7171932
- DOI: 10.1093/ijnp/pyz071
Amygdala-Based Altered miRNome and Epigenetic Contribution of miR-128-3p in Conferring Susceptibility to Depression-Like Behavior via Wnt Signaling
Abstract
Background: Recent studies suggest that microRNAs (miRNAs) can participate in depression pathogenesis by altering a host of genes that are critical in corticolimbic functioning. The present study focuses on examining whether alterations in the miRNA network in the amygdala are associated with susceptibility or resiliency to develop depression-like behavior in rats.
Methods: Amygdala-specific altered miRNA transcriptomics were determined in a rat depression model following next-generation sequencing method. Target prediction analyses (cis- and trans) and qPCR-based assays were performed to decipher the functional role of altered miRNAs. miRNA-specific target interaction was determined using in vitro transfection assay in neuroblastoma cell line. miRNA-specific findings from the rat in vivo model were further replicated in postmortem amygdala of major depressive disorder (MDD) subjects.
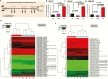
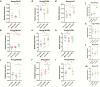
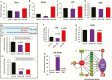
Results: Changes in miRNome identified 17 significantly upregulated and 8 significantly downregulated miRNAs in amygdala of learned helpless (LH) compared with nonlearned helpless rats. Prediction analysis showed that the majority of the upregulated miRNAs had target genes enriched for the Wnt signaling pathway. Among altered miRNAs, upregulated miR-128-3p was identified as a top hit based on statistical significance and magnitude of change in LH rats. Target validation showed significant downregulation of Wnt signaling genes in amygdala of LH rats. A discernable increase in expression of amygdalar miR-128-3p along with significant downregulation of key target genes from Wnt signaling (WNT5B, DVL, and LEF1) was noted in MDD subjects. Overexpression of miR-128-3p in a cellular model lead to a marked decrease in the expression of Dvl1 and Lef1 genes, confirming them as validated targets of miR-128-3p. Additional evidence suggested that the amygdala-specific diminished expression of transcriptional repressor Snai1 could be potentially linked to induced miR-128-2 expression in LH rats. Furthermore, an amygdala-specific posttranscriptional switching mechanism could be active between miR-128-3p and RNA binding protein Arpp21 to gain control over their target genes such as Lef1.
Conclusion: Our study suggests that in amygdala a specific set of miRNAs may play an important role in depression susceptibility, which could potentially be mediated through Wnt signaling.
Keywords: Wnt signaling; amygdala; depression; epigenetics; miRNA; stress.
© The Author(s) 2020. Published by Oxford University Press on behalf of CINP.
Figures


Comment in
-
Commentary on the study of Roy et al. Amygdala Based Altered mir-128-3p in Conferring Susceptibility to Depression-like Behavior via Wnt Signalling.Int J Neuropsychopharmacol. 2020 Apr 21;23(3):178-180. doi: 10.1093/ijnp/pyaa011. Int J Neuropsychopharmacol. 2020. PMID: 32072167 Free PMC article.
References
-
- Balakathiresan NS, Chandran R, Bhomia M, Jia M, Li H, Maheshwari RK (2014) Serum and amygdala microRNA signatures of posttraumatic stress: fear correlation and biomarker potential. J Psychiatr Res 57:65–73. - PubMed
-
- Belzeaux R, Bergon A, Jeanjean V, Loriod B, Formisano-Tréziny C, Verrier L, Loundou A, Baumstarck-Barrau K, Boyer L, Gall V, Gabert J, Nguyen C, Azorin JM, Naudin J, Ibrahim EC (2012) Responder and nonresponder patients exhibit different peripheral transcriptional signatures during major depressive episode. Transl Psychiatry 2:e185. - PMC - PubMed
-
- Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, Corrada D, Milanesi L, Gennarelli M (2013) Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol 23:602–611. - PubMed
-
- Chou HY, Howng SL, Cheng TS, Hsiao YL, Lieu AS, Loh JK, Hwang SL, Lin CC, Hsu CM, Wang C, Lee CI, Lu PJ, Chou CK, Huang CY, Hong YR (2006) GSKIP is homologous to the Axin GSK3beta interaction domain and functions as a negative regulator of GSK3beta. Biochemistry 45:11379–11389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

