Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma
- PMID: 32174248
- PMCID: PMC7549682
- DOI: 10.1080/15476286.2020.1741282
Using RNA sequencing to identify a putative lncRNA-associated ceRNA network in laryngeal squamous cell carcinoma
Abstract
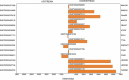
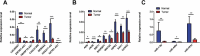
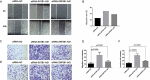
Accumulating evidence indicates that lncRNAs can interact with miRNAs to regulate target mRNAs through competitive interactions. However, this mechanism remains largely unexplored in laryngeal squamous cell carcinoma (LSCC). In this study, transcriptome-wide RNA sequencing was performed on 3 pairs of LSCC tissues and adjacent normal tissues to investigate the expression profiles of lncRNAs, miRNAs and mRNAs, with differential expression of 171 lncRNAs, 36 miRNAs and 1709 mRNAs detected. Seven lncRNAs, eight mRNAs and three miRNAs were identified to be dysregulated in patients' tissues by using qRT-PCR. GO and KEGG pathway enrichment analyses were performed to elucidate the potential functions of these differentially expressed genes in LSCC. Subsequently, a ceRNA (lncRNA-miRNA-mRNA) network including 4631 ceRNA pairs was constructed based on predicted miRNAs shared by lncRNAs and mRNAs. Cis- and transregulatory lncRNAs were analysed by bioinformatics-based methods. Importantly, mRNA-related ceRNA networks (mRCNs) were further obtained based on potential cancer-related coding genes. Coexpression between lncRNAs and downstream mRNAs was used as a criterion for the validation of mRCNs, with the ZNF561-AS1-miR217-WNT5A and SATB1-AS1-miR1299-SAV1/CCNG2/SH3 KBP1/JADE1/HIPK2 ceRNA regulatory interactions determined, followed by experimental validation after siRNA transfection. Moreover, ceRNA activity analysis revealed that different activities of ceRNA modules existing in specific pathological environments may contribute to the tumorigenesis of LSCC. Consistently, both downregulated SATB1-AS1 and ZNF561-AS1 significantly promoted laryngeal cancer cell migration and invasion, indicating their important roles in LSCC via a ceRNA regulatory mechanism. Taken together, the results of this investigation uncovered and systemically characterized a lncRNA-related ceRNA regulatory network that may be valuable for the diagnosis and treatment of LSCC.
Keywords: CeRNA; RNA sequencing; laryngeal carcinoma; lncRNA; noncoding RNA.
Conflict of interest statement
The authors declare that there is no conflict of interest associated with this study.
Figures






References
-
- Steuer CE, El-Deiry M, Parks JR, et al. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. - PubMed
-
- Groome P, O’Sullivan B, Irish J, et al. Management and outcome differences in supraglottic cancer between Ontario, Canada, and the surveillance, epidemiology, and end results areas of the United States. J Clin Oncol. 2003;21(3):496–505. - PubMed
-
- Gioacchini FM, Alicandri-Ciufelli M, Magliulo G, et al. The clinical relevance of Ki-67 expression in laryngeal squamous cell carcinoma. Eur Arch Oto-rhino-laryngol. 2015;272(7):1569–1576. - PubMed
-
- Lionello M, Staffieri A, Marioni G.. Potential prognostic and therapeutic role for angiogenesis markers in laryngeal carcinoma. Acta Otolaryngol. 2012;132(6):574–582. - PubMed
-
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
