Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 From Arabidopsis thaliana
- PMID: 32174940
- PMCID: PMC7056826
- DOI: 10.3389/fpls.2020.00157
Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 From Arabidopsis thaliana
Erratum in
-
Corrigendum: Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 From Arabidopsis thaliana.Front Plant Sci. 2021 Dec 23;12:826504. doi: 10.3389/fpls.2021.826504. eCollection 2021. Front Plant Sci. 2021. PMID: 35003199 Free PMC article.
Abstract
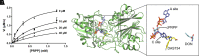
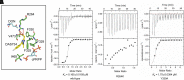
Chloroplastic glutamine phosphoribosylpyrophosphate amidotransferase (GPRATase) catalyzes the first committed step of de novo purine biosynthesis in Arabidopsis thaliana, and DAS734 is a direct and specific inhibitor of AtGPRAT, with phytotoxic effects similar to the leaf beaching phenotypes of known AtGPRAT genetic mutants, especially cia1 and atd2. However, the structure of AtGPRAT and the inhibition mode of DAS734 still remain poorly understood. In this study, we solved the structure of AtGPRAT2, which revealed structural differences between AtGPRAT2 and bacterial enzymes. Kinetics assay demonstrated that DAS734 behaves as a competitive inhibitor for the substrate phosphoribosyl pyrophosphate (PRPP) of AtGPRAT2. Docking studies showed that DAS734 forms electrostatic interactions with R264 and hydrophobic interactions with several residues, which was verified by binding assays. Collectively, our study provides important insights into the inhibition mechanism of DAS734 to AtGPRAT2 and sheds light on future studies into further development of more potent herbicides targeting Arabidopsis GPRATases.
Keywords: Arabidopsis thaliana; DAS734; X-ray crystallography; chloroplastic glutamine phosphoribosylpyrophosphate amidotransferase; competitive inhibition; herbicide.
Copyright © 2020 Cao, Du, Han, Zhou, Ren, Wang, Chen and Zhang.
Figures


Similar articles
-
Chemical genetic identification of glutamine phosphoribosylpyrophosphate amidotransferase as the target for a novel bleaching herbicide in Arabidopsis.Plant Physiol. 2007 Jul;144(3):1292-304. doi: 10.1104/pp.107.099705. Epub 2007 Jun 1. Plant Physiol. 2007. PMID: 17616508 Free PMC article.
-
Corrigendum: Crystal Structure of the Chloroplastic Glutamine Phosphoribosylpyrophosphate Amidotransferase GPRAT2 From Arabidopsis thaliana.Front Plant Sci. 2021 Dec 23;12:826504. doi: 10.3389/fpls.2021.826504. eCollection 2021. Front Plant Sci. 2021. PMID: 35003199 Free PMC article.
-
5-Phosphoribosylpyrophosphate amidotransferase from soybean root nodules: kinetic and regulatory properties.Arch Biochem Biophys. 1984 Mar;229(2):623-31. doi: 10.1016/0003-9861(84)90195-4. Arch Biochem Biophys. 1984. PMID: 6538402
-
Gout and the regulation of purine biosynthesis.Horiz Biochem Biophys. 1976;2:134-62. Horiz Biochem Biophys. 1976. PMID: 776767 Review.
-
The amidotransferase family of enzymes: molecular machines for the production and delivery of ammonia.Biochemistry. 1999 Jun 22;38(25):7891-9. doi: 10.1021/bi990871p. Biochemistry. 1999. PMID: 10387030 Review.
Cited by
-
Iron-sulfur clusters as inhibitors and catalysts of viral replication.Nat Chem. 2022 Mar;14(3):253-266. doi: 10.1038/s41557-021-00882-0. Epub 2022 Feb 14. Nat Chem. 2022. PMID: 35165425 Review.
-
A first-in-class dimethyl 2-acetamido terephthalate inhibitor targeting Conyza canadensis SHMT1 with a novel herbicidal mode-of-action.J Adv Res. 2024 Aug;62:59-70. doi: 10.1016/j.jare.2023.10.003. Epub 2023 Oct 10. J Adv Res. 2024. PMID: 37820886 Free PMC article.
-
Conformational Changes of Glutamine 5'-Phosphoribosylpyrophosphate Amidotransferase for Two Substrates Analogue Binding: Insight from Conventional Molecular Dynamics and Accelerated Molecular Dynamics Simulations.Front Chem. 2021 Feb 26;9:640994. doi: 10.3389/fchem.2021.640994. eCollection 2021. Front Chem. 2021. PMID: 33718330 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

