Comparison of Long Non-Coding RNA Expression Profiles of Cattle and Buffalo Differing in Muscle Characteristics
- PMID: 32174968
- PMCID: PMC7054449
- DOI: 10.3389/fgene.2020.00098
Comparison of Long Non-Coding RNA Expression Profiles of Cattle and Buffalo Differing in Muscle Characteristics
Abstract
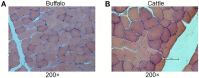
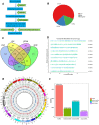
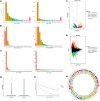
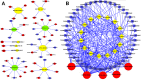
Buffalo meat consist good qualitative characteristics as it contains "thined tender" which is favorable for cardavascular system. However, the regulatory mechanisms of long non-coding RNA (lncRNA), differences in meat quality are not well known. The chemical-physical parameters revealed the muscle quality of buffalo that can be equivalent of cattle, but there are significant differences in shearing force and muscle fiber structure. Then, we examined lncRNA expression profiles of buffalo and cattle skeletal muscle that provide first insights into their potential roles in buffalo myogenesis. Here, we profiled the expression of lncRNA in cattle and buffalo skeletal muscle tissues, and 16,236 lncRNA candidates were detected with 865 up-regulated lncRNAs and 1,296 down-regulated lncRNAs when comparing buffalo to cattle muscle tissue. We constructed coexpression and ceRNA networks, and found lncRNA MSTRG.48330.7, MSTRG.30030.4, and MSTRG.203788.46 could be as competitive endogenous RNA (ceRNA) containing potential binding sites for miR-1/206 and miR-133a. Tissue expression analysis showed that MSTRG.48330.7, MSTRG.30030.4, and MSTRG.203788.46 were highly and specifically expressed in muscle tissue. Present study may be used as a reference tool for starting point investigations into the roles played by several of those lncRNAs during buffalo myogenesis.
Keywords: RNA-seq; buffalo; cattle; lncRNA; myogenesis.
Copyright © 2020 Li, Huang, Wang, Feng, Shi, Cui, Luo, Shafique, Qian, Ruan and Liu.
Figures

References
LinkOut - more resources
Full Text Sources

