High-Throughput Sequencing With the Preselection of Markers Is a Good Alternative to SNP Chips for Genomic Prediction in Broilers
- PMID: 32174971
- PMCID: PMC7056902
- DOI: 10.3389/fgene.2020.00108
High-Throughput Sequencing With the Preselection of Markers Is a Good Alternative to SNP Chips for Genomic Prediction in Broilers
Abstract
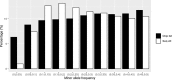
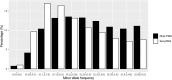
The choice of a genetic marker genotyping platform is important for genomic prediction in livestock and poultry. High-throughput sequencing can produce more genetic markers, but the genotype quality is lower than that obtained with single nucleotide polymorphism (SNP) chips. The aim of this study was to compare the accuracy of genomic prediction between high-throughput sequencing and SNP chips in broilers. In this study, we developed a new SNP marker screening method, the pre-marker-selection (PMS) method, to determine whether an SNP marker can be used for genomic prediction. We also compared a method which preselection marker based results from genome-wide association studies (GWAS). With the two methods, we analysed body weight at the12th week (BW) and feed conversion ratio (FCR) in a local broiler population. A total of 395 birds were selected from the F2 generation of the population, and 10X specific-locus amplified fragment sequencing (SLAF-seq) and the Illumina Chicken 60K SNP Beadchip were used for genotyping. The genomic best linear unbiased prediction method (GBLUP) was used to predict the genomic breeding values. The accuracy of genomic prediction was validated by the leave-one-out cross-validation method. Without SNP marker screening, the accuracies of the genomic estimated breeding value (GEBV) of BW and FCR were 0.509 and 0.249, respectively, when using SLAF-seq, and the accuracies were 0.516 and 0.232, respectively, when using the SNP chip. With SNP marker screening by the PMS method, the accuracies of GEBV of the two traits were 0.671 and 0.499, respectively, when using SLAF-seq, and 0.605 and 0.422, respectively, when using the SNP chip. Our SNP marker screening method led to an increase of prediction accuracy by 0.089-0.250. With SNP marker screening by the GWAS method, the accuracies of genomic prediction for the two traits were also improved, but the gains of accuracy were less than the gains with PMS method for all traits. The results from this study indicate that our PMS method can improve the accuracy of GEBV, and that more accurate genomic prediction can be obtained from an increased number of genomic markers when using high-throughput sequencing in local broiler populations. Due to its lower genotyping cost, high-throughput sequencing could be a good alternative to SNP chips for genomic prediction in breeding programmes of local broiler populations.
Keywords: chickens; feed conversion ratio; genomic prediction; high-throughput sequencing; marker screening method.
Copyright © 2020 Liu, Luo, Ma, Wang, Shu, Su and Qu.
Figures

References
-
- Allen D. M. (1971). Mean square error of prediction as a criterion for selecting variables. Technometrics 13, 469–475. 10.1080/00401706.1971.10488811 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

