Tumor slice culture as a biologic surrogate of human cancer
- PMID: 32175407
- PMCID: PMC7049013
- DOI: 10.21037/atm.2019.12.88
Tumor slice culture as a biologic surrogate of human cancer
Abstract
Background: The tumor microenvironment (TME) is critical to every aspect of cancer biology. Organotypic tumor slice cultures (TSCs) preserve the original TME and have demonstrated utility in predicting drug sensitivity, but the association between clinicopathologic parameters and in vitro TSC behavior has not been well-defined.
Methods: One hundred and eight fresh tumor specimens from liver resections at a tertiary academic center were procured and precisely cut with a Vibratome to create 250 μm × 6 mm slices. These fixed-dimension TSCs were grown on polytetrafluoroethylene inserts, and their metabolic activities were determined by a colorimetric assay. Correlation between baseline activities and clinicopathologic parameters was assessed. Tissue CEA mRNA expression was determined by RNAseq.
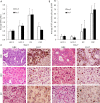
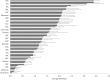
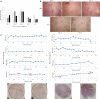
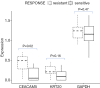
Results: By standardizing the dimensions of a slice, we found that adjacent tumor slices have equivalent metabolic activities, while those derived from different tumors exhibit >30-fold range in baseline MTS absorbances, which correlated significantly with the percentage of tumor necrosis based on histologic assessment. Extending this to individual cancers, we were able to detect intra-tumoral heterogeneity over a span of a few millimeters, which reflects differences in tumor cell density and Ki-67 positivity. For colorectal cancers, tissue CEA expression based on RNAseq of tumor slices was found to correlate with clinical response to chemotherapies.
Conclusions: We report a standardized method to assess and compare human cancer growth ex vivo across a wide spectrum of tumor samples. TSC reflects the state of tumor behavior and heterogeneity, thus providing a simple approach to study of human cancers with an intact TME.
Keywords: Organotypic; cancer model; heterogeneity; microenvironment.
2020 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: The authors have no conflicts of interest to declare.
Figures

References
LinkOut - more resources
Full Text Sources
