Variant calling on the GRCh38 assembly with the data from phase three of the 1000 Genomes Project
- PMID: 32175479
- PMCID: PMC7059836
- DOI: 10.12688/wellcomeopenres.15126.2
Variant calling on the GRCh38 assembly with the data from phase three of the 1000 Genomes Project
Abstract
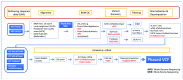
We present a set of biallelic SNVs and INDELs, from 2,548 samples spanning 26 populations from the 1000 Genomes Project, called de novo on GRCh38. We believe this will be a useful reference resource for those using GRCh38. It represents an improvement over the "lift-overs" of the 1000 Genomes Project data that have been available to date by encompassing all of the GRCh38 primary assembly autosomes and pseudo-autosomal regions, including novel, medically relevant loci. Here, we describe how the data set was created and benchmark our call set against that produced by the final phase of the 1000 Genomes Project on GRCh37 and the lift-over of that data to GRCh38.
Keywords: Genomics; population genetics; single nucleotide variation; variant calling; variant discovery.
Copyright: © 2019 Lowy-Gallego E et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

