Pim1 maintains telomere length in mouse cardiomyocytes by inhibiting TGFβ signalling
- PMID: 32176281
- PMCID: PMC7797214
- DOI: 10.1093/cvr/cvaa066
Pim1 maintains telomere length in mouse cardiomyocytes by inhibiting TGFβ signalling
Abstract
Aims: Telomere attrition in cardiomyocytes is associated with decreased contractility, cellular senescence, and up-regulation of proapoptotic transcription factors. Pim1 is a cardioprotective kinase that antagonizes the aging phenotype of cardiomyocytes and delays cellular senescence by maintaining telomere length, but the mechanism remains unknown. Another pathway responsible for regulating telomere length is the transforming growth factor beta (TGFβ) signalling pathway where inhibiting TGFβ signalling maintains telomere length. The relationship between Pim1 and TGFβ has not been explored. This study delineates the mechanism of telomere length regulation by the interplay between Pim1 and components of TGFβ signalling pathways in proliferating A549 cells and post-mitotic cardiomyocytes.
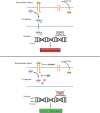
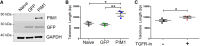
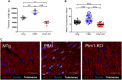
Methods and results: Telomere length was maintained by lentiviral-mediated overexpression of PIM1 and inhibition of TGFβ signalling in A549 cells. Telomere length maintenance was further demonstrated in isolated cardiomyocytes from mice with cardiac-specific overexpression of PIM1 and by pharmacological inhibition of TGFβ signalling. Mechanistically, Pim1 inhibited phosphorylation of Smad2, preventing its translocation into the nucleus and repressing expression of TGFβ pathway genes.
Conclusion: Pim1 maintains telomere lengths in cardiomyocytes by inhibiting phosphorylation of the TGFβ pathway downstream effectors Smad2 and Smad3, which prevents repression of telomerase reverse transcriptase. Findings from this study demonstrate a novel mechanism of telomere length maintenance and provide a potential target for preserving cardiac function.
Keywords: Cardiomyocyte; Pim1; Smad2; TGFβ; Telomere.
Published on behalf of the European Society of Cardiology. All rights reserved. © The Author(s) 2020. For permissions, please email: journals.permissions@oup.com.
Figures




Comment in
-
A novel signalling mechanism regulating telomere length in cardiomyocytes.Cardiovasc Res. 2021 Jan 1;117(1):13-14. doi: 10.1093/cvr/cvaa210. Cardiovasc Res. 2021. PMID: 32666074 No abstract available.

